Estimate a Generalized Linear Model (GLM).
Syntax
eq_name.glm(options) spec
List the glm keyword, followed by the dependent variable and a list of the explanatory variables, or an explicit linear expression.
If you enter an explicit linear specification such as “Y=C(1)+C(2)*X”, the response variable will be taken to be the variable on the left-hand side of the equality (“Y”) and the linear predictor will be taken from the right-hand side of the expression (“C(1)+C(2)*X”).
Offsets may be entered directly in an explicit linear expression or they may be entered as using the “offset=” option.
Specification Options
family=arg (default=“normal”) | Distribution family: Normal (“normal”), Poisson (“poisson”), Binomial Count (“binomial”), Binomial Proportion (“binprop”), Negative Binomial (“negbin”), Gamma (“gamma”), Inverse Gaussian (“igauss”), Exponential Mean (“emean)”, Power Mean (“pmean”), Binomial Squared (“binsq”). The Binomial Count, Binomial Proportion, Negative Binomial, and Power Mean families all require specification of a distribution parameter: |
n=arg (default=1) | Number of trials for Binomial Count (“family=binomial”) or Binomial Proportions (“family=binprop”) families. |
fparam=arg | Family parameter value for Negative Binomial (“family=negbin”) and Power Mean (“family=pmean”) families. |
link=arg (default=“identity”) | Link function: Identity (“identity”), Log (“log”), Log Compliment (“logc”), Logit (“logit”), Probit (“probit”), Log-log (“loglog”), Complementary Log-log (“cloglog”), Reciprocal (“recip”), Power (“power”), Box-Cox (“boxcox”), Power Odds Ratio (“opow”), Box-Cox Odds Ratio (“obox”). The Power, Box-Cox, Power Odds Ratio, and Box-Cox Odds Ratio links all require specification of a link parameter specified using “lparam=”. |
lparam=arg | Link parameter for Power (“link=power”), Box-Cox (“link=boxcox”), Power Odds Ratio (“link=opow”) and Box-Cox Odds Ratio (“link=obox”) link functions. |
offset=arg | Offset terms. |
disp=arg | Dispersion estimator: Pearson 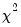 statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”). The default is family specific: “unit” for Binomial Count, Binomial Proportion, Negative Binomial, and Poison, and “pearson” for all others. The “deviance” option is only offered for families in the exponential family of distributions (Normal, Poisson, Binomial Count, Binomial Proportion, Negative Binomial, Gamma, Inverse Gaussian). |
dispval=arg | User-dispersion value (if “disp=user”). |
fwgts=arg | Frequency weights. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
In addition to the specification options, there are options for estimation and covariance calculation.
Additional Options
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “fisher” (IRLS – Fisher Scoring), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
estmeth=arg (default=”marquardt”) | Legacy estimation algorithm: Quadratic Hill Climbing (“marquardt”), Newton-Raphson (“newton”), IRLS - Fisher Scoring (“irls”), BHHH (“bhhh”). (Applicable when “optmethod=legacy”.) |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
preiter=arg (default=0) | Number of IRLS pre-iterations to refine starting values (only available for non-IRLS estimation). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “glm” (GLM method), “cr” (cluster robust). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “fisher” (expected Hessian). (Applicable when “optmethod=” not equal to “legacy”. |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate.(For non-cluster robust methods). |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). Applicable where “cov=hac”. |
covinfosel=arg (default=”aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). For settings where “cov=hac, covlag=a”. |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 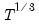 . For settings where “cov=hac, covlag=a”. |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). For settings where “cov=hac”. |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). For settings where “cov=hac” and “covkern=” is specified. |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). For settings where “cov=hac” and “covkern=” is specified. |
covbwoffset=number | Apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
covbwint | Use integer portion of kernel bandwidth. For settings where “cov=hac” and “covkern=” is specified. |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eqstrike.glm(link=log) numb c ip feb
estimates a normal regression model with exponential mean.
equation eqbinom.glm(family=binomial, n=total) disease c snore
estimates a binomial count model with default logit link where TOTAL contains the number of binomial trials and DISEASE is the number of binomial successes. The specification
equation eqbinom.glm(family=binprop, n=total, cov=huber, nodf) disease/total c snore
estimates the same specification in proportion form, and computes the coefficient covariance using the Huber-White sandwich with no d.f. correction.
equation eqprate.glm(family=binprop, disp=pearson) prate mprate log(totemp) log(totemp)^2 age age^2 sole
estimates a binomial proportions model with default logit link, but computes the coefficient covariance using the GLM scaled covariance with dispersion computed using the Pearson Chi-square statistic.
equation eqprate.glm(family=binprop, link=probit, cov=huber) prate mprate log(totemp) log(totemp)^2 age age^2 sole
estimates the same basic specification, but with a probit link and Huber-White standard errors.
equation testeq.glm(family=poisson, offset=log(pyears)) los hmo white type2 type3 c
estimates a Poisson specification with an offset term LOG(PYEARS).
Cross-references
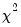 statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”).
statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”).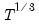 .
.