@rotmat factor rotation matrix: 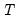 .
.
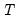 .
.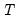 .
.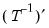 .
.anticov |
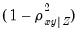 , where
, where 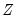 are the remaining variables.
are the remaining variables. p | Print the matrix. |
clearhist |
clearremarks |
copy |
display |
displayname |
eigen |
source=arg (default= “observed”) | Source matrix to be analyzed: “observed” (observed covariance matrix), “scaled” (scaled observed matrix), “reducedinit” (reduced using initial uniquenesses), “reduced” (reduced using final uniquenesses). |
eigvec | Add the eigenvectors to the table of eigenvalue results. May be combined with the “matrix” keyword. |
matrix | Display the source matrix along with the table of eigenvalue results. May be combined with the “eigvec” keyword. |
scree | Display eigenvalue graph of the ordered eigenvalues (Scree plot). May be combined with the “diff” and “cproport” keywords. |
diff | Display graph of the difference in successive eigenvalues. May be combined with the “scree” and “cproport” keywords. |
cproport | Display graph of the cumulative proportion of total variance associated with each eigenvalue/eigenvector. May be combined with the “scree” and “diff” keywords. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
factnames |
factor |
fitstats |
p | Print the results. |
fitted |
common | Display common covariance.(default is to display the fitted covariance). |
p | Print the matrix. |
fsel |
p | Print the results. |
gls |
rescale | Rescale the uniqueness and loadings estimates so that they match the observed variances. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
n=arg or fsmethod=arg (default=“map”) | Number of factors: “kaiser” (Kaiser-Guttman greater than mean), “mineigen” (Minimum eigenvalue criterion; specified using “eiglimit”), “varfrac” (fraction of variance accounted for; specified using “varlimit”), “map” (Velicer’s Minimum Average Partial method), “bstick” (comparison with broken stick distribution), “parallel” (parallel analysis: number of replications specified using “pnreps”; “pquant” indicates the quantile method value if employed), “scree” (standard error scree method), “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), integer (user-specified integer value). |
eiglimit=number (default=1) | Limit value for retaining factors using the eigenvalue comparison (where “n=mineigen”). |
varlimit=number (default=0.5) | Fraction of total variance explained limit for retaining factors using the variance limit criterion (where “n=varlimit”). |
porig | Use the unreduced matrix for parallel analysis (the default is to use the reduced matrix). For parallel analysis only (“n=parallel”). |
preps= integer (default=100) | Number of parallel analysis repetitions. For parallel analysis only (“n=parallel”). |
pquant=number | Quantile value for parallel analysis comparison (if not specified, the mean value will be employed). For parallel analysis only (“n=parallel”). |
pseed=positive integer | Seed the random number generator for parallel analysis. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. For parallel analysis only (“n=parallel”). |
Type of random number generator for the simulation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). For parallel analysis only (“n=parallel”). | |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 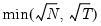 ), “size” ( ), “size” (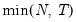 ), “user” (user specified value), where ), “user” (user specified value), where 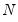 is the number of series and is the number of series and 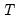 is the number of observations. is the number of observations.(1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let 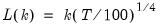 for 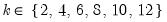 and let and let 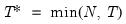 ; then the default maximum lag is given by ; then the default maximum lag is given by 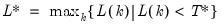 |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
priors=arg | Method for obtaining initial communalities: “smc” (squared multiple correlations), “max” (maximum absolute correlation”), “pace” (noniterative partitioned covariance estimation), “frac” (fraction of the diagonals of the original matrix; specified using “priorfrac=”), “random” (random fractions of the original diagonals), “user” (user-specified vector; specified using “priorunique”). |
priorfrac=number | User-specified common fraction (between 0 and 1) to be used when “priors=frac”. |
priorunique=arg | Vector of initial uniqueness estimates to be used when “priors=user”. By default, the values will be taken from the corresponding elements of the coefficient vector C. |
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). User-specified covariances are indicated by specifying a sym matrix object in place of a list of series or groups in the command. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
ipf |
heywood=arg (default=“stop”) | Method for handling Heywood cases (negative uniqueness estimates): “stop” (stop and report final results), “last” (stop and report previous iteration results”, “reset” (set negative uniquenesses to zero and continue), “ignore” (ignore and continue). |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
n=arg or fsmethod=arg (default=“map”) | Number of factors: “kaiser” (Kaiser-Guttman greater than mean), “mineigen” (Minimum eigenvalue criterion; specified using “eiglimit”), “varfrac” (fraction of variance accounted for; specified using “varlimit”), “map” (Velicer’s Minimum Average Partial method), “bstick” (comparison with broken stick distribution), “parallel” (parallel analysis: number of replications specified using “pnreps”; “pquant” indicates the quantile method value if employed), “scree” (standard error scree method), “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), integer (user-specified integer value). |
eiglimit=number (default=1) | Limit value for retaining factors using the eigenvalue comparison (where “n=mineigen”). |
varlimit=number (default=0.5) | Fraction of total variance explained limit for retaining factors using the variance limit criterion (where “n=varlimit”). |
porig | Use the unreduced matrix for parallel analysis (the default is to use the reduced matrix). For parallel analysis only (“n=parallel”). |
preps= integer (default=100) | Number of parallel analysis repetitions. For parallel analysis only (“n=parallel”). |
pquant=number | Quantile value for parallel analysis comparison (if not specified, the mean value will be employed). For parallel analysis only (“n=parallel”). |
pseed=positive integer | Seed the random number generator for parallel analysis. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. For parallel analysis only (“n=parallel”). |
Type of random number generator for the simulation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). For parallel analysis only (“n=parallel”). | |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 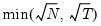 ), “size” ( ), “size” (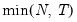 ), “user” (user specified value), where ), “user” (user specified value), where 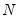 is the number of series and is the number of series and 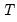 is the number of observations. is the number of observations.(1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let 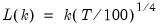 for 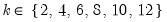 and let and let 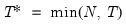 ; then the default maximum lag is given by ; then the default maximum lag is given by 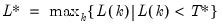 |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
priors=arg | Method for obtaining initial communalities: “smc” (squared multiple correlations), “max” (maximum absolute correlation”), “pace” (noniterative partitioned covariance estimation), “frac” (fraction of the diagonals of the original matrix; specified using “priorfrac=”), “random” (random fractions of the original diagonals), “user” (user-specified vector; specified using “priorunique”). |
priorfrac=number | User-specified common fraction (between 0 and 1) to be used when “priors=frac”. |
priorunique=arg | Vector of initial uniqueness estimates to be used when “priors=user”. By default, the values will be taken from the corresponding elements of the coefficient vector C. |
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). User-specified covariances are indicated by specifying a sym matrix object in place of a list of series or groups in the command. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
label |
c | Clears all text fields in the label. |
d | Sets the description field to text. |
s | Sets the source field to text. |
u | Sets the units field to text. |
r | Appends text to the remarks field as an additional line. |
p | Print the label view. |
loadings |
graph | Display graphs of the loadings (default is to display the loadings in a spreadsheet view). |
unrotated | Use the unrotated loadings (default is to use the rotated loadings, if available). |
prompt | Force the dialog to appear from within a program (for loadings graphs only) |
p | Print results. |
mult =arg (default=“first”) | Multiple series handling: plot first against remainder (“first”), plot as x-y pairs (“pair”), lower-triangular plot (“lt”). |
nocenter | Do not center graphs around the origin. By default, EViews centers biplots around (0, 0). |
makescores |
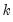 factor names, EViews will save the first
factor names, EViews will save the first 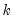 factors to the workfile. The factors will be named using the specified list, appended with the name modifiers, if specified.
factors to the workfile. The factors will be named using the specified list, appended with the name modifiers, if specified.unrotated | Use unrotated loadings in computations (the default is to use the rotated loadings, if available). |
type =arg (default=“exact”) | Exact coefficient (“exact”), coarse adjusted factor coefficients (“coefs”), coarse adjusted factor loadings (“loadings”). |
coef=arg (default=“reg”) | Method for computing the factor score coefficient matrix: Thurstone regression (“reg”), Ideal Variables (“ideal”), Bartlett weighted least squares (“wls”), generalized Anderson-Rubin-McDonald (“anderson”), Green (“green”). For “type=exact” and “type=coefs” specifications. |
coarse=arg (default=“unrestrict”) | Method for computing the coarse (-1, 0, 1) scores coefficients (Grice, 1991a): Unrestricted -- (“unrestrict”) coef weights set based only on sign; Unique–recode (“recode”) only element with highest value is coded to a non-zero value; Unique–drop (“drop”) only elements with loadings not in excess of the threshold are set to non-zero values. For “type=coefs” and “type=loadings” specifications. |
cutoff=number (default = 0.3) | Cutoff value for coarse score coefficient calculation (Grice, 1991a). For “type=coef” specifications, the cutoff value represents the fraction of the largest absolute coefficient weight per factor against which the absolute exact score coefficients should be compared. For “type=loadings”, and “type=struct” specifications, the cutoff is the value against which the absolute loadings or structure coefficients should be compared. |
moment=arg (default =“est”; if feasible) | Standardize the observables data using means and variances from: original estimation (“est”), or the computed moments from specified observable variables (“obs”). The “moment=est” option is only available for factor models estimated using Pearson or uncentered Pearson correlation and covariances since the remaining models involve unobserved or non-comparable moments. |
df | Degrees-of-freedom correct the observables variances computed when “moment=obs” (divide sums-of-squares by 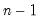 instead of instead of 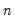 ). ). |
n=arg | (Optional) Name of group object to contain the factor score series. |
coefout | (Optional) Name of matrix in which to save the factor score coefficient matrix. |
prompt | Force the dialog to appear from within a program. |
maxcor |
p | Print the matrix. |
rescale | Rescale the uniqueness and loadings estimates so that they match the observed variances. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
n=arg or fsmethod=arg (default=“map”) | Number of factors: “kaiser” (Kaiser-Guttman greater than mean), “mineigen” (Minimum eigenvalue criterion; specified using “eiglimit”), “varfrac” (fraction of variance accounted for; specified using “varlimit”), “map” (Velicer’s Minimum Average Partial method), “bstick” (comparison with broken stick distribution), “parallel” (parallel analysis: number of replications specified using “pnreps”; “pquant” indicates the quantile method value if employed), “scree” (standard error scree method), “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), integer (user-specified integer value). |
eiglimit=number (default=1) | Limit value for retaining factors using the eigenvalue comparison (where “n=mineigen”). |
varlimit=number (default=0.5) | Fraction of total variance explained limit for retaining factors using the variance limit criterion (where “n=varlimit”). |
porig | Use the unreduced matrix for parallel analysis (the default is to use the reduced matrix). For parallel analysis only (“n=parallel”). |
preps= integer (default=100) | Number of parallel analysis repetitions. For parallel analysis only (“n=parallel”). |
pquant=number | Quantile value for parallel analysis comparison (if not specified, the mean value will be employed). For parallel analysis only (“n=parallel”). |
pseed=positive integer | Seed the random number generator for parallel analysis. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. For parallel analysis only (“n=parallel”). |
Type of random number generator for the simulation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). For parallel analysis only (“n=parallel”). | |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 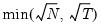 ), “size” ( ), “size” (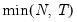 ), “user” (user specified value), where ), “user” (user specified value), where 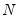 is the number of series and is the number of series and 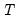 is the number of observations. is the number of observations.(1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let 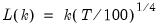 for 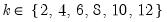 and let and let 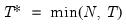 ; then the default maximum lag is given by ; then the default maximum lag is given by 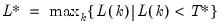 |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
priors=arg | Method for obtaining initial communalities: “smc” (squared multiple correlations), “max” (maximum absolute correlation”), “pace” (noniterative partitioned covariance estimation), “frac” (fraction of the diagonals of the original matrix; specified using “priorfrac=”), “random” (random fractions of the original diagonals), “user” (user-specified vector; specified using “priorunique”). |
priorfrac=number | User-specified common fraction (between 0 and 1) to be used when “priors=frac”. |
priorunique=arg | Vector of initial uniqueness estimates to be used when “priors=user”. By default, the values will be taken from the corresponding elements of the coefficient vector C. |
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). User-specified covariances are indicated by specifying a sym matrix object in place of a list of series or groups in the command. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
msa |
p | Print the results. |
observed |
scaled | Scale the observed matrix so that it has unit diagonals. |
p | Print the results. |
olepush |
output |
p | Print view. |
pace |
rescale | Rescale the uniqueness and loadings estimates so that they match the observed variances. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
n=arg or fsmethod=arg (default=“map”) | Number of factors: “kaiser” (Kaiser-Guttman greater than mean), “mineigen” (Minimum eigenvalue criterion; specified using “eiglimit”), “varfrac” (fraction of variance accounted for; specified using “varlimit”), “map” (Velicer’s Minimum Average Partial method), “bstick” (comparison with broken stick distribution), “parallel” (parallel analysis: number of replications specified using “pnreps”; “pquant” indicates the quantile method value if employed), “scree” (standard error scree method), “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), integer (user-specified integer value). |
eiglimit=number (default=1) | Limit value for retaining factors using the eigenvalue comparison (where “n=mineigen”). |
varlimit=number (default=0.5) | Fraction of total variance explained limit for retaining factors using the variance limit criterion (where “n=varlimit”). |
porig | Use the unreduced matrix for parallel analysis (the default is to use the reduced matrix). For parallel analysis only (“n=parallel”). |
preps= integer (default=100) | Number of parallel analysis repetitions. For parallel analysis only (“n=parallel”). |
pquant=number | Quantile value for parallel analysis comparison (if not specified, the mean value will be employed). For parallel analysis only (“n=parallel”). |
pseed=positive integer | Seed the random number generator for parallel analysis. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. For parallel analysis only (“n=parallel”). |
Type of random number generator for the simulation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). For parallel analysis only (“n=parallel”). | |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 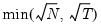 ), “size” ( ), “size” (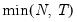 ), “user” (user specified value), where ), “user” (user specified value), where 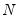 is the number of series and is the number of series and 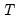 is the number of observations. is the number of observations.(1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let 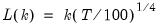 for 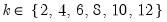 and let and let 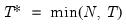 ; then the default maximum lag is given by ; then the default maximum lag is given by 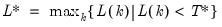 |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). User-specified covariances are indicated by specifying a sym matrix object in place of a list of series or groups in the command. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
partcor |
p | Print the matrix. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
n=arg or fsmethod=arg (default=“map”) | Number of factors: “kaiser” (Kaiser-Guttman greater than mean), “mineigen” (Minimum eigenvalue criterion; specified using “eiglimit”), “varfrac” (fraction of variance accounted for; specified using “varlimit”), “map” (Velicer’s Minimum Average Partial method), “bstick” (comparison with broken stick distribution), “parallel” (parallel analysis: number of replications specified using “pnreps”; “pquant” indicates the quantile method value if employed), “scree” (standard error scree method), “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), integer (user-specified integer value). |
eiglimit=number (default=1) | Limit value for retaining factors using the eigenvalue comparison (where “n=mineigen”). |
varlimit=number (default=0.5) | Fraction of total variance explained limit for retaining factors using the variance limit criterion (where “n=varlimit”). |
porig | Use the unreduced matrix for parallel analysis (the default is to use the reduced matrix). For parallel analysis only (“n=parallel”). |
preps= integer (default=100) | Number of parallel analysis repetitions. For parallel analysis only (“n=parallel”). |
pquant=number | Quantile value for parallel analysis comparison (if not specified, the mean value will be employed). For parallel analysis only (“n=parallel”). |
pseed=positive integer | Seed the random number generator for parallel analysis. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. For parallel analysis only (“n=parallel”). |
Type of random number generator for the simulation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). For parallel analysis only (“n=parallel”). | |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 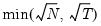 ), “size” ( ), “size” (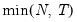 ), “user” (user specified value), where ), “user” (user specified value), where 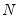 is the number of series and is the number of series and 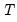 is the number of observations. is the number of observations.(1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let 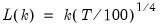 for 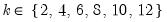 and let and let 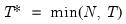 ; then the default maximum lag is given by ; then the default maximum lag is given by 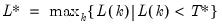 |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
priors=arg | Method for obtaining initial communalities: “smc” (squared multiple correlations), “max” (maximum absolute correlation”), “pace” (noniterative partitioned covariance estimation), “frac” (fraction of the diagonals of the original matrix; specified using “priorfrac=”), “random” (random fractions of the original diagonals), “user” (user-specified vector; specified using “priorunique”). |
priorfrac=number | User-specified common fraction (between 0 and 1) to be used when “priors=frac”. |
priorunique=arg | Vector of initial uniqueness estimates to be used when “priors=user”. By default, the values will be taken from the corresponding elements of the coefficient vector C. |
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). User-specified covariances are indicated by specifying a sym matrix object in place of a list of series or groups in the command. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
reduced |
initial | Display the reduced matrix computed using the initial uniqueness estimates. |
p | Print the matrix. |
resids |
common | Display the residuals computed using only the common fitted covariance. |
p | Print the matrix. |
rotate |
type=arg (default=“orthog”) | Orthogonal (“orthog”) or oblique (“oblique”) rotation (ignored if method is not supported, e.g, “orthogonal Harris-Kaiser” or “oblique Entropy Ratio”). |
method=arg (default=“varimax”) | Method (objective) for the rotation. See keywords below |
param=arg | Rotation parameter, if applicable (see description below). |
preparam=arg (default=1, Varimax) | Orthomax pre-rotation parameter (for “method=hk” and “method=promax”). |
Method | Keyword | Orthogonal | Oblique |
Biquartimax | biquartimax | • | • |
Crawford-Ferguson | cf | • | • |
Entropy | entropy | • | |
Entropy Ratio | entratio | • | |
Equamax | equamax | • | • |
Factor Parsimony | parsimony | • | • |
Generalized Crawford-Ferguson | gcf | • | • |
Geomin | geomin | • | • |
Harris-Kaiser (case II) | hk | • | |
Infomax | infomax | • | • |
Oblimax | oblimax | • | |
Oblimin | oblimin | • | |
Orthomax | orthomax | • | • |
Parsimax | parsimax | • | • |
Pattern Simplicity | pattern | • | • |
Promax | promax | • | |
Quartimax/Quartimin | quartimax | • | • |
Simplimax | simplimax | • | • |
Tandem I | tandemi | • | |
Tandem II | tandemii | • | |
Target | target | • | • |
Varimax | varimax | • | • |
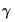 is evaluated using the Crawford-Ferguson objective with factor complexity weight
is evaluated using the Crawford-Ferguson objective with factor complexity weight 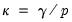 (see
“Types of Rotation”).
(see
“Types of Rotation”). 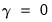 ), Varimax (
), Varimax (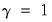 ), Equamax (
), Equamax (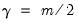 ), Parsimax (
), Parsimax (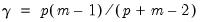 ) and Factor Parsimony (
) and Factor Parsimony (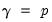 ).
).Method | 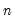 | Parameter Description |
Crawford-Ferguson | 1 | Factor complexity weight. The variable complexity weight is 1 minus the factor complexity weight. (default=0, Quartimax) |
Generalized Crawford-Ferguson | 4 | Vector of weights for (in order): total squares, variable complexity, factor complexity, diagonal quartics. (no default) |
Geomin | 1 | Epsilon offset. (default=0.01) |
Harris-Kaiser (case II) | 2 | Power parameter (default=0, independent cluster solution), Orthomax pre-rotation parameter. (default=1, Varimax) |
Oblimin | 1 | Deviation from orthogonality. (default=0, Quartimin) |
Orthomax | 1 | Factor complexity weight. (default=1, Varimax) |
Promax | 2 | Power parameter (default=3), Orthomax pre-rotation parameter (default=1, Varimax). |
Simplimax | 1 | Fraction of near-zero loadings. (default=0.75) |
Target | 1 | Name of 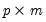 matrix of target loadings. Missing values correspond to unrestricted elements. matrix of target loadings. Missing values correspond to unrestricted elements.(no default) |
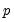 is the number of variables and
is the number of variables and 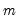 is the number of factors. The remaining options modify the properties of the specified rotation method:
is the number of factors. The remaining options modify the properties of the specified rotation method:wgts=arg (default=“none”) | Row weighting for loadings: none (“none”), kaiser (“kaiser”), Cureton-Mulaik (“cureton”). |
prior=arg (default =“unrotated”) | Initial rotation matrix: unrotated (“unrotated”), randomly generated (“random”), previous rotation (“previous”), user-specified (“user”). |
ptype=arg (default=“orthog”) | Type of prior random rotation: orthogonal (“orthog”) or oblique (“oblique”). Only relevant if “prior=random” and the main rotation method is oblique. If the main rotation method is orthogonal, random prior rotations will be orthogonalized. |
preps=integer (default=25) | Number of random prior rotations to evaluate (maximum 10000). |
pname=arg | Name of matrix containing prior rotation. |
pseed=positive integer | Seed the random number generator for the prior random rotations. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. |
Type of random number generator for the random prior rotation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). | |
m=integer | Maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the norm of the gradients scaled by the objective function. The criterion will be set to the nearest value between 1e-24 and 0.2. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
p | Print rotation results. |
rotateclear |
rotateout |
p | Print the table of results. |
setattr |
scores |
out=arg (default=“table”) | Output format: coefficient summary and diagnostics (“table”), spreadsheet table of scores (“sheet”), graph of scores (“graph”), graph of scores with loadings axes (“biplot”). |
unrotated | Use unrotated loadings in computations (the default is to use the rotated loadings, if available). |
type =arg (default=“exact”) | Exact coefficient (“exact”), coarse adjusted factor coefficients (“coefs”), coarse adjusted factor loadings (“loadings”). |
coef=arg (default=“reg”) | Method for computing the exact or coarse adjusted factor score coefficient matrix: Thurstone regression (“reg”), Ideal Variables (“ideal”), Bartlett weighted least squares (“wls”), generalized Anderson-Rubin-McDonald (“anderson”), Green (“green”). For “type=exact” and “type=coefs” specifications. |
coarse=arg (default=“unrestrict”) | Method for computing the coarse (-1, 0, 1) scores coefficients (Grice, 1991a): Unrestricted -- (“unrestrict”) coef weights set based only on sign; Unique–recode (“recode”) only element with highest value is coded to a non-zero value; Unique–drop (“drop”) only elements with loadings not in excess of the threshold are set to non-zero values. For “type=coefs” and “type=loadings” specifications. |
cutoff=number (default = 0.3) | Cutoff value for coarse scores coefficient calculations (Grice, 1991a). For “type=coefs” specifications, the cutoff value represents the fraction of the largest absolute coefficient weight per factor against which the exact score coefficients should be compared. For “type=loadings” specifications, the cutoff is the value against which the absolute loadings or structure coefficients should be compared. |
moment=arg (default =“est”; if feasible) | Standardize the observables data using means and variances from: original estimation (“est”), the computed moments from specified observable variables (“obs”). The “moment=est” option is only available for factor models estimated using Pearson or uncentered Pearson correlation and covariances since the remaining models involve unobserved or non-comparable moments. |
df | Degrees-of-freedom correct the observables variances computed when “moment=obs” (divide sums-of-squares by 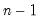 instead of instead of 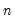 ). ). |
coefout | (Optional) Name of matrix in which to save factor score coefficient matrix. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
mult =arg (default=“first”) | Multiple series handling for graphs: plot first against remainder (“first”), plot as x-y pairs (“pair”), lower-triangular plot (“lt”) |
nocenter | Do not center graphs around the origin. |
labels=arg, (default=“outlier”) | Observation labels for scores: outliers only (“outlier”), all points (“all”), none (“none”). |
labelprob=number | Probability value for determining whether a point is an outlier according to the chi-square tests based on the squared Mahalanbois distance between the observation and the sample means (when using the “labels=outlier” option). |
userscale=arg | User-scale factor to be applied to the unscaled loadings (setting this option overrides the automatic scaling). |
autoscale=arg (default = 1) | User-scale factor to be applied to the automatic loadings scale (when displaying both loadings and scores). |
smc |
p | Print the matrix. |
structure |
p | Print the matrix. |
uls |
rescale | Rescale the uniqueness and loadings estimates so that they match the observed variances. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
n=arg or fsmethod=arg (default=“map”) | Number of factors: “kaiser” (Kaiser-Guttman greater than mean), “mineigen” (Minimum eigenvalue criterion; specified using “eiglimit”), “varfrac” (fraction of variance accounted for; specified using “varlimit”), “map” (Velicer’s Minimum Average Partial method), “bstick” (comparison with broken stick distribution), “parallel” (parallel analysis: number of replications specified using “pnreps”; “pquant” indicates the quantile method value if employed), “scree” (standard error scree method), “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), integer (user-specified integer value). |
eiglimit=number (default=1) | Limit value for retaining factors using the eigenvalue comparison (where “n=mineigen”). |
varlimit=number (default=0.5) | Fraction of total variance explained limit for retaining factors using the variance limit criterion (where “n=varlimit”). |
porig | Use the unreduced matrix for parallel analysis (the default is to use the reduced matrix). For parallel analysis only (“n=parallel”). |
preps= integer (default=100) | Number of parallel analysis repetitions. For parallel analysis only (“n=parallel”). |
pquant=number | Quantile value for parallel analysis comparison (if not specified, the mean value will be employed). For parallel analysis only (“n=parallel”). |
pseed=positive integer | Seed the random number generator for parallel analysis. If not specified, EViews will seed the random number generator with a single integer draw from the default global random number generator. For parallel analysis only (“n=parallel”). |
Type of random number generator for the simulation: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). For parallel analysis only (“n=parallel”). | |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 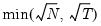 ), “size” ( ), “size” (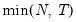 ), “user” (user specified value), where ), “user” (user specified value), where 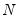 is the number of series and is the number of series and 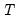 is the number of observations. is the number of observations.(1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let 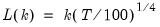 for 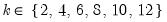 and let and let 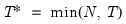 ; then the default maximum lag is given by ; then the default maximum lag is given by 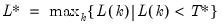 |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
priors=arg | Method for obtaining initial communalities: “smc” (squared multiple correlations), “max” (maximum absolute correlation”), “pace” (noniterative partitioned covariance estimation), “frac” (fraction of the diagonals of the original matrix; specified using “priorfrac=”), “random” (random fractions of the original diagonals), “user” (user-specified vector; specified using “priorunique”). |
priorfrac=number | User-specified common fraction (between 0 and 1) to be used when “priors=frac”. |
priorunique=arg | Vector of initial uniqueness estimates to be used when “priors=user”. By default, the values will be taken from the corresponding elements of the coefficient vector C. |
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). User-specified covariances are indicated by specifying a sym matrix object in place of a list of series or groups in the command. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |