Command Reference
Commands
The following list summarizes the EViews basic commands.
Other chapters document different aspects of the command language:
Command Actions
do execute action without opening window.
preview preview objects contained in a database or workfile.
Global Commands
cd change default directory.
exit exit the EViews program.
help displays the documentation.
output redirect printer output.
param set parameter values.
rndseed set the seed of the random number generator.
smpl set the sample range.
Object Creation Commands
data enter data from keyboard.
factor factor analysis object.
frml numeric or alpha series object with a formula for auto-updating.
genr numeric or alpha series object.
graph graph object—create using a graph command or by merging existing graphs.
link series or alpha link object.
var var estimation object.
Object Container, Data, and File Commands
ccopy copy series from DRI database.
cfetch fetch series from DRI database.
clabel display DRI series description.
close close object, program, or workfile.
copy copy objects within and between workfiles, workfile pages, and databases.
db open or create a database.
dbcopy make copy of a database.
delete delete objects from a workfile.
driconvert convert the entire DRI database to an EViews database.
fetch fetch objects from databases or databank files.
hconvert convert an entire Haver Analytics database to an EViews database.
hfetch fetch series from a Haver Analytics database.
hlabel obtain label from a Haver Analytics database.
import imports data from a foreign file or a previously saved workfile into the current default workfile.
importattr imports observation values stored inside one or more series in a second workfile page into the attribute fields of objects within the current workfile page.
importmat imports data from a foreign file into a matrix object in the current workfile.
importtbl imports data from a foreign file into a table object in the current workfile.
open open a program or text (ASCII) file.
optsave save the current EViews global options settings .INI files into a directory.
optset replace the current EViews global options settings .INI files with ones based in a different directory.
pageload load one or more pages into a workfile from a workfile or a foreign data source.
pagerefresh refresh all links and auto-series in the active workfile page—primarily used to refresh links that use external database data.
pagesave save page into a workfile or a foreign data source.
pagestack reshape the workfile page by stacking observations.
pagestruct apply a workfile structure to the page.
pageunlink break links in all link objects and auto-updating series (formulae) in the active workfile page.
pageunstack reshape the workfile page by unstacking observations into multiple series.
range reset the workfile range
.
read import data from a foreign disk file into series.
save save workfile to disk.
store store objects in database and databank files.
unlink break links and auto-updating series (formulae) in the specified series objects.
wfcompare compare the contents of the current workfile or page with the contents of a different workfile, page, or database.
wfdetails change the details displayed in the current workfile window.
wfdir change the workfile view to a simple object directory listing.
wffilter change the workfile object filter for the current workfile window.
wfopen open workfile or foreign source data as a workfile.
wforder change the workfile page order.
wfrefresh refresh all links and auto-series in the active workfile—primarily used to refresh links that use external database data.
wfsave save workfile to disk as a workfile or a foreign data source.
wfsnapshot takes a manual snapshot of the current workfile.
wfstats display the workfile statistics and summary view.
wfunlink break links in all link objects and auto-updating series (formulae) in the active workfile.
wfuse activate a workfile.
workfile create or change active workfile.
write write series to a disk file.
Object Utility Commands
close close window of an object, program, or workfile.
Object Assignment Commands
data enter data from keyboard.
frml assign formula for auto-updating to a numeric or alpha series object.
genr create numeric or alpha series object.
nrnd fill object with standard normal random numbers.
rmvnorm fill object with multivariate normal random numbers.
rnd fill object with uniform random numbers.
rndint fill object with random integers.
rndseed set random number generator seed.
Matrix Utility Commands
colplace Places column vector into matrix.
matplace Places matrix object in another matrix object.
mtos Converts a matrix object to series, alpha, or group.
nrnd Fill the matrix with normal random numbers .
rmvnorm Fill the matrix with multivariate normal random numbers.
rnd Fill the matrix with uniform random numbers.
rowplace Places a rowvector in matrix object.
stom Converts series, alpha, or group to vector or matrix after removing observations with NAs.
stomna Converts series, alpha or group to vector or matrix without removing observations with NAs.
ttom Fills a matrix with the numeric contents of a table.
Graph Creation Commands
Graph creation is discussed in detail in
“Graph Creation Command Summary”.
hilo high-low(-open-close) graph.
qqplot quantile-quantile graph.
scatmat matrix of all pairwise scatterplots.
Table Commands
setcell format and fill in a table cell.
setline place a horizontal line in table.
tabplace insert a table into another table.
ttom fills a matrix with the numeric contents of a table.
Note that with the exception of
tabplace, these commands are supported primarily for backward compatibility. There is a more extensive set of table procs for working with and customizing tables. See
“Table Procs”.
Programming Commands
addin register an Add-in.
adduo register a user object.
clearerrs sets the current program error count to 0.
commandcap send text to the command capture window.
deleteaddin unregister a program file as an EViews Add-in.
logclear clears the log window of a program.
logclose closes one or more or all message log windows.
logeval sends result of the command to a log window.
logmode sets logging of specified messages.
logmsg adds a line of text to the program log.
logsave saves the program log to a text file.
open open a program file.
optimize find the solution to a user-defined optimization problem.
output redirects print output to objects or files.
poff turns off automatic printing in programs.
pon turns on automatic printing in programs.
seterr sets a user-specified execution error.
seterrcount sets the current program execution error count.
setmaxerrs sets the maximum number of errors that a program may encounter before execution is halted.
spawn spawn a new process.
toc display elapsed time (since timer reset) in seconds.
External Interface Commands
xclose close an open connection to an external application.
xget retrieve data from an external application into an EViews object.
xlog switch on or off the external application log inside EViews.
xoff turns off mode that sends all subsequent command lines to the external program.
xon turns on mode that sends all subsequent command lines to the external program.
xopen open a connection to an external application.
xpackage installs the specified R package in the current external R connection.
xput send an EViews object to an external application.
xrun run a command in an external application.
Interactive Use Commands
The following commands have object command forms (e.g.
, Equation::arch). These commands are particularly suited for interactive command line use. In general, we recommend that you use the object forms of the commands.
arch estimate autoregressive conditional heteroskedasticity (ARCH and GARCH) models.
archtest LM test for the presence of ARCH in the residuals.
ardl autoregressive distributed lag models.
auto Breusch-Godfrey serial correlation Lagrange Multiplier (LM) test.
binary binary dependent variable models (includes probit, logit, gompit) models.
breakls least squares with breakpoints and breakpoint determination
cause pairwise Granger causality tests.
censored estimate censored and truncated regression (includes tobit) models.
chow Chow breakpoint and forecast tests for structural change.
coint cointegration test.
cointreg estimate cointegrating equation using FMOLS, CCR, or DOLS.
count count data modeling (includes poisson, negative binomial and quasi-maximum likelihood count models).
data enter data from keyboard.
did estimate a panel equation using the difference-in-difference estimator.
enet elastic net regression (including Lasso and ridge regression).
facbreak factor breakpoint test for stability.
factest estimate a factor analysis model.
fit static forecast from an equation.
forecast dynamic forecast from an equation.
funcoef estimate a functional coefficient regression equation.
glm estimate a Generalized Linear Model (GLM).
gmm estimate an equation using generalized method of moments.
heckit estimate a selection equation using the Heckman ML or 2-step method.
hist histogram and descriptive statistics.
hpf Hodrick-Prescott filter.
liml estimate an equation using Limited Information Maximum Likelihood and K-class Estimation.
logit logit (binary) estimation.
ls equation using least squares or nonlinear least squares.
midas estimate an equation using Mixed Data Sampling (MIDAS) regression.
ordered ordinal dependent variable models (includes ordered probit, ordered logit, and ordered extreme value models).
probit probit (binary) estimation.
qreg estimate an equation using quantile regression.
reset Ramsey’s RESET test for functional form.
robustls robust regression (M-estimation, S-estimation and MM-estimation).
seas seasonal adjustment for quarterly and monthly time series.
stats descriptive statistics.
switchreg exogenous and Markov switching regression.
testadd likelihood ratio test for adding variables to equation.
testdrop likelihood ratio test for dropping variables from equation.
threshold threshold least squares, including threshold autoregression and smooth threshold autoregression.
tsls estimate an equation using two-stage least squares regression.
varest specify and estimate a VAR or VEC.
varsel equation estimation using least squares with variable selection (uni-directional, stepwise, swapwise, combinatorial, Auto-GETS, Lasso).
Register a program file as an EViews Add-in.
Syntax
addin(options) [path\]prog_name
registers the specified program file as an EViews Add-in. Note that the program file should have a “.PRG” extension, which you need not specify in the prog_name.
If you do not provide the optional path specification, EViews looks for the program file in the default EViews Add-ins directory.
Explicit path specifications containing “.\” and “..\” (to indicate the current level and one directory level up) are evaluated relative the directory of the installer program in which the addin command is specified, or the EViews default directory if addin is run from the command line.
You may use the special “<addins>”directory keyword in your path specification.
Options
type=arg | Specify the Add-ins type, where arg is the name of a EViews object type. The default is to create a global Add-in. Specifying an object-specific Add-in using a matrix object as in “type=matrix”, “type=vector”, etc. will register the Add-in for all matrix object types (including coef, rowvector, and sym objects). Sample objects do not support object-specific Add-ins so that “type=sample” is not allowed. |
proc=arg | User--defined command/procedure name. If omitted, the Add-in will not have a command form. |
menu=arg | Text for the Add-in menu entry. If omitted, the Add-in will not have an associated menu item. Note that you may use the “&” symbol in the entry text to indicate that the following character should be used as a menu shortcut. |
desc=arg | Brief description of the Add-in that will be displayed in the Add-ins management dialog. |
docs=arg | Path and filename for the Add-in documentation. Determination of the path follows the rules specified above for the addin filename. |
version=arg | Version number of the Add-in. If no version number is supplied, EViews will assume version 1.0. |
url=arg | Specify the location of an XML file containing information on the Add-in used for updating the Add-in to the latest version. If not supplied, EViews will default to an XML file hosted on the EViews website. |
nowarn | Removes the prompt warning that an add-in already exists with the same name (and forces an overwrite of that add-in). |
Examples
addin(proc="myaddin", desc="This is my add-in", version="1.0") .\myaddin.prg
registers the file “Myaddin.prg” as a global Add-in, with the user-defined global command myaddin, no menu support, and no assigned documentation file. The description “This is my add-in” will appear in the main Add-ins management dialog. The version number is “1.0”. Note that the “.\” indicates the directory from which the program containing the addin command was run, or the EViews default directory if addin is run interactively.
addin(type="graph", menu="Add US Recession Shading", proc="recshade", docs=".\recession shade.txt", desc="Applies US recession shading to a graph object.") .\recshade.prg
registers the file “Recshade.prg” as a graph specific Add-in. The Add-in supports the object-command recshade, has an object-specific menu item “Add US Recession Shading”, and has a documentation file “Recession shade.txt”.
addin(type=equation, menu="Simple rolling regression", proc=roll, docs="<addins>\Roll\Roll.pdf", desc="Rolling Regression - simple version", url="www.mysite.com/myaddins.xml", version="1.2") "<addins>\Roll\roll.prg"
registers the Add-in file “Roll.prg” as an equation specific Add-in. Note that the documentation and program files are located in the “Roll” subdirectory of the default Add-ins directory. The XML file located at www.mysite.com/myaddins.xml is used when checking for available updates for the Add-in, and the current version number is set to “1.2”.
Cross-references
See
“Add-ins” for a detailed discussion of Add-ins.
Register an EViews user object class.
Syntax
adduo(options) [path\]definition_name
registers the specified definition file as an EViews user object. Note that the definition file should have a “.INI” extension.
If you do not provide the optional path specification, EViews looks for the program file in the default EViews user objects directory.
Explicit path specifications containing “.\” and “..\” (to indicate the current level and one directory level up) are evaluated relative the directory of the installer program in which the adduo command is specified, or the EViews default directory if adduo is run from the command line.
Options
name=arg | Specify the name of the user object class. |
desc=arg | Brief description of the user object that will be displayed in the user object management dialog. |
docs=arg | Path and filename for the user object documentation. Determination of the path follows the rules specified above for the adduo filename. |
version=arg | Version number of the Add-in. If no version number is supplied, EViews will assume version 1.0. |
url=arg | Specify the location of an XML file containing information on the Add-in used for updating the Add-in to the latest version. If not supplied, EViews will default to an XML file hosted on the EViews website. |
Examples
adduo(name="roll", desc="Rolling Regression Object") .\rolldef.ini
registers the roll class of user object, specifying a description of “Rolling Regression Object”, and using the definition file rolldef.ini, located in the same folder as the installer program.
adduo(name="resstore", version="1.0", url="www.mysite.com/myuos.xml") .\resstoredef.ini
registers the resstore class of user object, specifying the version number as “1.0”, and using the XML file located at “www.mysite.com/myuos.xml” to check for updates.
Cross-references
See
“User Objects” for a discussion of user objects.
Estimate generalized autoregressive conditional heteroskedasticity (GARCH) models.
Syntax
arch(p,q,options) y [x1 x2 x3] [@ p1 p2 [@ t1 t2]]
arch(p,q,options) y=expression [@ p1 p2 [@ t1 t2]]
The first two options specify the order of the GARCH model:
• The arch estimation method specifies a GARCH(p, q) model with p ARCH terms and q GARCH terms. Note the order of the arguments in which the ARCH and GARCH terms are entered.
The maximum value for
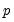
or
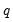
is 9; values above will be set to 9. The minimum value for
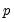
is 1. The minimum value for
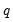
is 0. If either

or
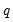
is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default.
• For CGARCH, FIEGARCH and MIDAS-GARCH models, EViews only estimates (1,1) models. For these specifications,
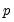
and
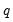
options should not be specified, and if provided, will be ignored.
After the “ARCH” keyword and options, specify the dependent variable followed by a list of regressors in the mean equation.
• By default, only the intercept is included in the conditional variance equation. If you wish to specify variance regressors, list them after the mean equation using an “@”-sign to separate the mean from the variance equation.
• When estimating component ARCH models, you may specify exogenous variance regressors for both the permanent and transitory components. After the mean equation regressors, first list the regressors for the permanent component, followed by an “@”-sign, then the regressors for the transitory component. A constant term is always included as a permanent component regressor.
• For MIDAS-GARCH models, the low-frequency permanent component regressor are entered after the mean equation regressors and an “@”-sign. The regressor should be specified as pagename\seriesname.
Options
Type Options
The default is to estimate a standard GARCH model. You may specify one of the followings keywords to estimate a different model:
egarch | Exponential GARCH. |
parch[=arg] | Power ARCH. If the optional arg is provided, the power parameter will be set to that value, otherwise the power parameter will be estimated. |
cgarch | Component (permanent and transitory) ARCH. |
figarch | Fractional GARCH (FIGARCH). |
fiegarch | Fractional Exponential GARCH (FIEGARCH(1,1)). |
midas | MIDAS GARCH(1,1) |
General Options
thrsh | For Component GARCH models, include a threshold term. |
thrsh=integer (default=0) | Number of threshold terms for GARCH models. The maximum number of terms allowed is 9. |
vt | Variance target of the constant term for GARCH models. (May not be used with integrated specifications.) |
integrated | Restrict GARCH model to be integrated, i.e. IGARCH. (May not be used with variance targeting.) |
asy=integer (default=1) | Number of asymmetric terms in Power ARCH or EGARCH models. The maximum number of terms allowed is 9. |
trunclag=integer (default=1000) | Number of terms in the expansion approximation for FIGARCH and FIEGARCH models. |
archm=arg | ARCH-M (ARCH in mean) specification with the conditional standard deviation (“archm=sd”), the conditional variance (“archm=var”), or the log of the conditional variance (“archm= log”) entered as a regressor in the mean equation. |
tdist [=number] | Estimate the model assuming that the residuals follow a conditional Student’s t-distribution (the default is the conditional normal distribution). Providing the optional number greater than two will fix the degrees of freedom to that value. If the argument is not provided, the degrees of freedom will be estimated. |
ged [=number] | Estimate the model assuming that the residuals follow a conditional GED (the default is the conditional normal distribution). Providing a positive value for the optional argument will fix the GED parameter. If the argument is not provided, the parameter will be estimated. |
z | Turn of backcasting for both initial MA innovations and initial variances. |
backcast=n | Backcast weight to calculate value used as the presample conditional variance. Weight needs to be greater than 0 and less than or equal to 1; the default value is 0.7. Note that a weight of 1 is equivalent to no backcasting, i.e. using the unconditional residual variance as the presample conditional variance. |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). “bfgs” is the default for new equations. |
optstep = arg | Step method: “marquardt” (Marquardt - default); “dogleg” (Dogleg); “linesearch” (Line search). (Applicable when “optmethod=bfgs”, “optmethod=newton” or “optmethod=opg”.) |
b | Use Berndt-Hall-Hall-Hausman (BHHH) as maximization algorithm. The default is Marquardt. (Applicable when “optmethod=legacy”.) |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “bollerslev” (Bollerslev-Wooldridge method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “ (Applicable when non-legacy “optmethod=” with “cov=ordinary”.) |
h | Bollerslev-Wooldridge robust quasi-maximum likelihood (QML) covariance/standard errors. (Applicable for “optmethod=legacy” when estimating assuming normal errors.) |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of preliminary LS estimates (out of range values are set to “s=1”). |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. Available only for legacy estimation (“optmeth=legacy”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
MIDAS Options
lag=arg | Specify the number of lags of the low frequency regressor to include. Default value is 32. |
beta=arg | Beta function restriction: none (“none”), trend coefficient equals 1 (“trend”), endpoints coefficient equals 0 (“end-point”), both trend and endpoints restriction (“both”). For use when “midwgt=beta”. The default is “beta=none”. |
thrsh | Include a threshold term. |
optmethod=arg | Optimization method for nonlinear estimation: “bfgs” (BFGS); “newton” Newton-Raphson), “opg”, “bhhh” (OPG or BHHH), or “hybrid” (initial BHHH followed by BFGS). Hybrid is the default method. |
optstep=arg | Step method for nonlinear estimation: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method for nonlinear models: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich). |
covinfo=arg | Information matrix method for nonlinear models: “opg” (OPG); “hessian” (observed Hessian). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values in nonlinear estimation. If the “s=number” or “s” options are not used, EViews will use random starting values. |
s=number | Determine starting values for nonlinear estimation. Specify a number between zero and oSpecify the number of lags of the low frequency regressor to include. Default value is 32.ne representing the fraction of preliminary EViews chosen values. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. If the “s=number” or “s” options are not used, EViews will use random starting values. |
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator used in random starting values. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
showopts/-showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector; the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
arch(4, 0, m=1000, cov=bollerslev) sp500 c
estimates an ARCH(4) model with a mean equation consisting of the series SP500 regressed on a constant. The procedure will perform up to 1000 iterations, and will report Bollerslev-Wooldridge robust QML standard errors upon completion.
The commands:
c = 0.1
arch(thrsh=1, s, mean=var) @pch(nys) c ar(1)
estimate a TARCH(1, 1)-in-mean specification with the mean equation relating the percent change of NYS to a constant, an AR term of order 1, and a conditional variance (GARCH) term. The first line sets the default coefficient vector to 0.1, and the “s” option uses these values as coefficient starting values.
The command:
arch(1, 2, asy=0, parch=1.5, ged=1.2) dlog(ibm)=c(1)+c(2)* dlog(sp500) @ r
estimates a symmetric Power ARCH(2, 1) (autoregressive GARCH of order 2, and moving average ARCH of order 1) model with GED errors. The power of model is fixed at 1.5 and the GED parameter is fixed at 1.2. The mean equation consists of the first log difference of IBM regressed on a constant and the first log difference of SP500. The conditional variance equation includes an exogenous regressor R.
Cross-references
See
“ARCH and GARCH Estimation” for a discussion of ARCH models.
See
Equation::arch for the equivalent object command.
Test for autoregressive conditional heteroskedasticity (ARCH).
Carries out Lagrange Multiplier (LM) tests for ARCH in the residuals.
Note that a more general version of the ARCH test is available using
Equation::archtest.
Syntax
archtest(options)
Options
You must specify the order of ARCH for which you wish to test. The number of lags to be included in the test equation should be provided in parentheses after the arch keyword.
Other Options:
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
ls output c labor capital
archtest(4)
Regresses OUTPUT on a constant, LABOR, and CAPITAL, and tests for ARCH up to order 4.
equation eq1.arch sp500 c
archtest(4)
Estimates a GARCH(1,1) model with mean equation of SP500 on a constant and tests for additional ARCH up to order 4. Note that when performing an archtest after an arch estimation, EViews uses the standardized residuals (the residual of the mean equation divided by the estimated conditional standard deviation) to form the test.
Cross-references
See
“ARCH LM Test” for further discussion of testing ARCH and
“ARCH and GARCH Estimation” for a discussion of working with ARCH models in EViews.
See
Equation::archtest for the equivalent object command. See
Equation::hettest for a more general version of the ARCH test.
Estimate an equation with autoregressive distributed lags using linear and nonlinear least squares or quantile regression.
Syntax
equation.ardl(options) linear_regs [@ static_regs] [@asy dual_asymmetric_regs] [@asylr long_run_asymmetric_regs] [@asysr short_run_asymmetric_regs]
The linear_regs specification is required:
• The linear_regs list should be the dependent variable followed by a list of linear distributed-lag regressors.
The remaining specifications are optional
• static_regs should be a list of static regressors, not including a constant or trend term.
• dual_asymmetric_regs are distributed-lag regressors which are asymmetric both in the short-run and long-run.
• long_run_asymmetric_regs regressors are distributed lag-regressors which are asymmetric in the long-run but symmetric in the short-run.
• short_run_asymmetric_regs are asymmetric regressors which are distributed lag-regressors which are asymmetric in the short-run but symmetric in the long-run.
You may specify the lag for an individual distributed-lag variable using the “@fl(variable, lag)” syntax. For instance, if the variable X should use 3 lags, irrespective of the fixed or automatic lag settings, you may specify this by entering “@fl(x, 3)” in the regressor list.
Options
Least Squares ARDL Options
method=arg (default = “ls”) | Set the method of estimation: "ls" (least-squares regression, default) or "qreg" (quantile regression). |
determ=arg (default = “rconst”) | Johansen deterministic trend type: “none” (no deterministics), “rconst” (restricted constant and no trend), “uconst” (unrestricted constant and no trend), “rtrend” (unrestricted constant and restricted trend, “utrend” (unrestricted constant and unrestricted trend). |
trend=arg (deprecated) | Johansen deterministic trend type: “none” (no deterministics), “const” (restricted constant and no trend, default), “uconst” (unrestricted constant and no trend), “linear” (unrestricted constant and restricted trend, “ulinear” (unrestricted constant and unrestricted trend). Note: this is a deprecated s option which handles a subset of cases covered by the “determ=” option |
fixed | Do not use automatic selection for lag lengths. This option must be used with the “deplags=” and “reglags=” options. |
deplags=int (default = 4) | Set the number of lags for the dependent variable to int. If automatic selection is used, this sets the maximum number of possible lags. If fixed lags are used (the fixed option is set), this fixes the number of lags. |
reglags=int (default = 4) | Set the number of lags for the explanatory variables (dynamic regressors) to int. If automatic selection is used, this sets the maximum number of possible lags. If fixed lags are used (the fixed option is set), this fixes the number of lags for each regressor. |
ic=key (default =“aic”) | Set the method of automatic model selection. key may take values of “aic” (Akaike information criterion, default), “bic” (Schwarz criterion), “hq” (Hannan-Quinn criterion) or “rbar2” (Adjusted R-squared, not applicable in panel workfiles). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “hac” (Newey-West HAC, available for nonlinear least squares or ARMA estimated by CLS).. |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of  . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Quantile ARDL Options
quant=number (default = 0.5) | Quantile to be fit (where number is a value between 0 and 1). |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=arg (default=“sandwich”) | Method for computing coefficient covariance matrix: “iid” (ordinary estimates), “sandwich” (Huber sandwich estimates), “boot” (bootstrap estimates). When “cov=iid” or “cov=sandwich”, EViews will use the sparsity nuisance parameter calculation specified in “spmethod=” when estimating the coefficient covariance matrix. |
bwmethod=arg (default = “hs”) | Method for automatically selecting bandwidth value for use in estimation of sparsity and coefficient covariance matrix: “hs” (Hall-Sheather), “bf” (Bofinger), “c” (Chamberlain). |
bw =number | Use user-specified bandwidth value in place of automatic method specified in “bwmethod=”. |
bwsize=number (default = 0.05) | Size parameter for use in computation of bandwidth (used when “bw=hs” and “bw=bf”). |
spmethod=arg (default=“kernel”) | Sparsity estimation method: “resid” (Siddiqui using residuals), “fitted” (Siddiqui using fitted quantiles at mean values of regressors), “kernel” (Kernel density using residuals) Note: “spmethod=resid” is not available when “cov=sandwich”. |
btmethod=arg (default= “pair”) | Bootstrap method: “resid” (residual bootstrap), “pair” (xy-pair bootstrap), “mcmb” (MCMB bootstrap), “mcmba” (MCMB-A bootstrap). |
btreps=integer (default=100) | Number of bootstrap repetitions |
btseed=positive integer | Seed the bootstrap random number generator. If not specified, EViews will seed the bootstrap random number generator with a single integer draw from the default global random number generator. |
btrnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator for the bootstrap: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
btobs=integer | Number of observations for bootstrap subsampling (when “bsmethod=pair”). Should be significantly greater than the number of regressors and less than or equal to the number of observations used in estimation. EViews will automatically restrict values to the range from the number of regressors and the number of estimation observations. If omitted, the bootstrap will use the number of observations used in estimation. |
btout=name | (optional) Matrix to hold results of bootstrap simulations. |
k=arg (default=“e”) | Kernel function for sparsity and coefficient covariance matrix estimation (when “spmethod=kernel”): “e” (Epanechnikov), “r” (Triangular), “u” (Uniform), “n” (Normal–Gaussian), “b” (Biweight–Quartic), “t” (Triweight), “c” (Cosinus). |
m=integer | Maximum number of iterations. |
s | Use the current coefficient values in estimator coefficient vector as starting values (see also
param). |
s=number (default =0) | Determine starting values for equations. Specify a number between 0 and 1 representing the fraction of preliminary least squares coefficient estimates. Note that out of range values are set to the default. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
opens example data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000.
The following command
ardl(deplags=8, reglags=8) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable, and the log of real GDP as a dynamic regressor. Quarterly dummy variables are included as static regressors. Automatic model selection is used to determine the number of lags of LOG(REALCONS) and LOG(REALGDP).
The command
ardl(deplags=3, reglags=3, fixed) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
estimates a second model, replicating Example 20.4 from Greene, with a fixed three lags of the dependent variable and three lags of the regressor.
ardl(deplags=1, reglags=1, fixed) log(realcons) log(realgdp) @asy log(realgovt)
The line above estimates an ARDL(1,1,1) model with the log of real consumption as the dependent variable, the log of real GDP as a linear regressor, and log of real government expenditures as a dual asymmetric regressor.
ardl(deplags=1, reglags=1, fixed) log(realcons) log(realgdp) @asy log(realgovt) @asysr log(realinvs)
extends the previous model and estimates an ARDL(1,1,1,1) model by including the log of real investments as a long-run asymmetric regressor.
ardl(deplags=1, reglags=1, fixed) log(realcons) log(realgdp) @asy log(realgovt) @asysr log(realinvs) @asylr log(tbilrate)
The line above extends the previous model and estimates an ARDL(1,1,1,1,1) model by including the log of treasury bill rates as a short-run asymmetric regressor.
wfopen oecd.wf1
ardl(fixed, deplags=1, reglags=1) log(cons) log(inf) log(inc)
This example estimates a panel ARDL model using the workfile “OECD.wf1”. This model replicates that given in the original Pesaran, Shin and Smith 1999 paper. Model selection is not used to choose the optimal lag lengths, rather a fixed single lag of both the dependent variable and the regressor is employed.
ardl(method=qreg, ls=fixed, deplags=1, reglags=1, quant=0.4) log(realcons) log(realgdp)
This command estimates a QARDL(1,1) model where lag selection is fixed for both the dependent and independent regressors, and the quantile value is 0.4.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
Compute serial correlation LM (Lagrange multiplier) test.
Carries out Breusch-Godfrey Lagrange Multiplier (LM) tests for serial correlation in the estimation residuals from the default equation.
Syntax
auto(order, options)
You must specify the order of serial correlation for which you wish to test. You should specify the number of lags in parentheses after the auto keyword, followed by any additional options.
Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
To regress OUTPUT on a constant, LABOR, and CAPITAL, and test for serial correlation of up to order four you may use the commands:
ls output c labor capital
auto(4)
The commands:
output(t) c:\result\artest.txt
equation eq1.ls cons c y y(-1)
auto(12, p)
perform a regression of CONS on a constant, Y and lagged Y, and test for serial correlation of up to order twelve. The first line redirects printed tables/text to the “Artest.TXT” file.
Cross-references
See
“Serial Correlation LM Test” for further discussion of the Breusch-Godfrey test.
See
Equation::auto for the corresponding equation view.
Estimate binary dependent variable models.
Estimates models where the binary dependent variable Y is either zero or one (probit, logit, gompit).
Syntax
binary(options) y x1 [x2 x3 ...]
binary(options) specification
Options
d=arg (default=“n”) | Specify likelihood: normal likelihood function, probit (“n”), logistic likelihood function, logit (“l”), Type I extreme value likelihood function, Gompit (“x”). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “glm” (GLM method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian - default). (Applicable when non-legacy “optmethod=”.) |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option applicable when “optmethod=legacy”). |
g | GLM standard errors and covariances. (Legacy option applicable when “optmethod=legacy”). |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
To estimate a logit model of Y using a constant, WAGE, EDU, and KIDS, and computing Huber-White standard errors, you may use the command:
binary(d=l,cov=huber) y c wage edu kids
Note that this estimation uses the default global optimization options. The commands:
param c(1) .1 c(2) .1 c(3) .1
binary(s) y c x2 x3
estimate a probit model of Y on a constant, X2, and X3, using the specified starting values. The commands:
coef beta_probit = @coefs
matrix cov_probit = @coefcov
store the estimated coefficients and coefficient covariances in the coefficient vector BETA_PROBIT and matrix COV_PROBIT.
Cross-references
See
Equation::binary for the corresponding equation method.
Estimation by linear least squares regression with breakpoints.
Syntax
breakls(options) y z1 [z2 z3 ...] [@nv x1 x2 x3 ...]
List the dependent variable first, followed by a list of the independent variables that have coefficients which are allowed to vary across breaks, followed optionally by the keyword @nv and a list of non-varying coefficient variables.
Options
Breakpoint Options
method=arg (default=“seqplus1”) | Breakpoint selection method: “seqplus1” (sequential tests of single 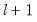 versus 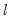 breaks), “seqall” (sequential test of all possible 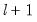 versus  breaks), “glob” (tests of global 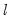 vs. no breaks), “globplus1” (tests of 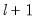 versus 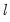 globally determined breaks), “globinfo” (information criteria evaluation), “user” (user-specified break dates). |
select=arg | Sub-method setting (options depend on “method=”). (1) if “method=glob”: Sequential (“seq”) (default), Highest significant (“high”), 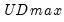 (“udmax”), 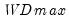 (“wdmax”). (2) if “method=globinfo”: Schwarz criterion (“bic” or “sic”) (default), Liu-Wu-Zidek criterion (“lwz”). |
trim=arg (default=5) | Trimming percentage for determining minimum segment size (5, 10, 15, 20, 25). |
maxbreaks=integer (default=5) | Maximum number of breakpoints to allow (not applicable if “method=seqall”). |
maxlevels=integer (default=5) | Maximum number of break levels to consider in sequential testing (applicable when “method=sequall”). |
breaks="arg" | User-specified break dates entered in double quotes. For use when “method=user”. |
size=arg (default=5) | Test sizes for use in sequential determination and final test evaluation (10, 5, 2.5, 1) corresponding to 0.10, 0.05, 0.025, 0.01, respectively |
heterr | Assume regimes specific error distributions in variance computation. |
commondata | Assume a common distribution for the data across segments (only applicable if original equation is estimated with a robust covariance method, “heterr” is not specified). |
General Options
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=keyword | Covariance type (optional): “white” (White diagonal matrix), “hac” (Newey-West HAC). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 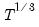 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
covbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
coef=arg | Specify the name of the coefficient vector; the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
breakls m1 c unemp
uses the Bai-Perron sequential
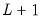
versus
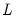
tests to determine the optimal breaks in a model regressing M1 on the breaking variables C and UNEMP.
breakls(method=glob, select=high) m1 c unemp
uses the global Bai-Perron
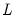
versus none tests to determine the breaks. The selected break will be the highest significant number of breaks.
breakls(size=5, trim=10) m1 c unemp
lowers the sequential test size from 0.10 to 0.05, and raises the trimming to 10 percent.
breakls(method=user, break=”1990q1 2010q4”) m1 c @nv unemp
estimates the model with two user-specified break dates. In addition, the variable UNEMP is restricted to have common coefficients across the regimes.
Cross-reference
See
Equation::multibreak for estimation of regression equations with breaks.
Granger causality test.
Performs pairwise Granger causality tests between (all possible) pairs of the listed series or group of series.
Syntax
cause(n, options) ser1 ser2 ser3
Following the keyword, list the series or group of series for which you wish to test for Granger causality.
Options
You must specify the number of lags n to use for the test by providing an integer in parentheses after the keyword. Note that the regressors of the test equation are a constant and the specified lags of the pair of series under test.
Other options:
prompt | Force tcointehe dialog to appear from within a program. |
p | Print output of the test. |
Examples
To compute Granger causality tests of whether GDP Granger causes M1 and whether M1 Granger causes GDP, you may enter the command:
cause(4) gdp m1
The regressors of each test are a constant and four lags of GDP and M1.
cause(12,p) m1 gdp r
prints the result of six pairwise Granger causality tests for the three series. The regressors of each test are a constant and twelve lags of the two series under test (and do not include lagged values of the third series).
Cross-references
See
“Granger Causality” for a discussion of Granger’s approach to testing hypotheses about causality.
See also
Group::cause for the corresponding group view.
Copy one or more series from the DRI Basic Economics database to EViews data bank (.DB) files.
You must have the DRI database installed on your computer to use this feature.
Syntax
ccopy series_name
Type the name of the series or wildcard expression for series you want to copy after the ccopy keyword. The data bank files will be stored in the default directory with the same name as the series names in the DRI database. You can supply path information to indicate the directory for the data bank files.
Examples
The command:
ccopy lhur
copies the DRI series LHUR to “Lhur.DB” file in the default path directory.
ccopy b:gdp c:\nipadata\gnet
copies the GDP series to the “Gdp.DB” file in the “b:” drive and the GNET series to the “Gnet.DB” file in “c:\nipadata”.
Cross-references
Change default directory.
The cd command changes the current default working directory. The current working directory is displayed in the “Path=...” message in the bottom right of the EViews window.
Note that the default directory is not used for processing of include files (see
include).
Syntax
cd path_name
Examples
To change the default directory to “sample data” in the “a:” drive, you may issue the command:
cd "a:\sample data"
Notice that the directory name is surrounded by double quotes. If your name does not contain spaces, you may omit the quotes. The command:
cd a:\test
changes the default directory to “a:\test”.
cd “<myonedrive>:\test
changes the default directory to the cloud location, MYONEDRIVE.
Subsequent save operations will save into the default directory, unless you specify a different directory at the time of the operation.
Cross-references
See
“Workfile Basics” for further discussion of basic operations in EViews.
Estimation of censored and truncated models.
Estimates models where the dependent variable is either censored or truncated. The allowable specifications include the standard Tobit model.
Syntax
censored(options) y x1 [x2 x3]
censored(options) specification
Options
l=number (default=0) | Set value for the left censoring limit. |
r=number (default=none) | Set value for the right censoring limit. |
l=series_name, i | Set series name of the indicator variable for the left censoring limit. |
r=series_name, i | Set series name of the indicator variable for the right censoring limit. |
t | Estimate truncated model. |
d=arg (default=“n”) | Specify error distribution: normal (“n”), logistic (“l”), Type I extreme value (“x”). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian - default). (Applicable when non-legacy “optmethod=”). |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
The command:
censored(cov=huber) hours c wage edu kids
estimates a censored regression model of HOURS on a constant, WAGE, EDU, and KIDS with QML standard errors. This command uses the default normal likelihood, with left-censoring at HOURS=0, no right censoring, and the quadratic hill climbing algorithm.
Cross-references
See
“Discrete and Limited Dependent Variable Models” for discussion of censored and truncated regression models.
See
Equation::censored for the corresponding equation method.
Fetch a series from the DRI Basic Economics database into a workfile.
cfetch reads one or more series from the DRI Basic Economics Database into the active workfile. You must have the DRI database installed on your computer to use this feature.
Syntax
cfetch series_name
Examples
cfetch lhur gdp gnet
reads the DRI series LHUR, GDP, and GNET into the current active workfile, performing frequency conversions if necessary.
Cross-references
EViews’ automatic frequency conversion is described in
“Frequency Conversion”.
Change default directory.
Chow test for stability.
Carries out Chow breakpoint or Chow forecast tests for parameter constancy.
Syntax
chow(options) obs1 [obs2 obs3 ...] @ x1 x2 x3
You must provide the breakpoint observations (using dates or observation numbers) to be tested. To specify more than one breakpoint, separate the breakpoints by a space. For the Chow breakpoint test, if the equation is specified by list and contains no linear terms, you may specify a subset of the regressors to be tested for a breakpoint after an “@” sign.
Options
f | Chow forecast test. For this option, you must specify a single breakpoint to test (default performs breakpoint test). |
p | Print the result of test. |
Examples
The commands:
ls m1 c gdp cpi ar(1)
chow 1970Q1 1980Q1
perform a regression of M1 on a constant, GDP, and CPI with first order autoregressive errors, and employ a Chow breakpoint test to determine whether the parameters before the 1970’s, during the 1970’s, and after the 1970’s are “stable”.
To regress the log of SPOT on a constant, the log of P_US, and the log of P_UK, and to carry out the Chow forecast test starting from 1973, enter the commands:
ls log(spot) c log(p_us) log(p_uk)
chow(f) 1973
To test whether only the constant term and the coefficient on the log of P_US prior to and after 1970 are “stable” enter the commands:
chow 1970 @ c log(p_us)
Cross-references
See
“Chow's Breakpoint Test” for further discussion.
Display a DRI Basic Economics database series description.
clabel reads the description of a series from the DRI Basic Economics Database and displays it in the status line at the bottom of the EViews window.
Use this command to verify the contents of a given DRI database series name. You must have the DRI database installed on your computer to use this feature.
Syntax
clabel series_name
Examples
clabel lhur
displays the description of the DRI series LHUR on the status line.
Cross-references
Set the current error count to 0.
May only be used in programs.
Close object, program, or workfile.
Closing an object eliminates its window. If the object is named, it is still displayed in the workfile as an icon, otherwise it is deleted. Closing a program or workfile eliminates its window and removes it from memory. If a workfile has changed since you activated it, you will see a dialog box asking if you want to save it to disk.
Syntax
close option_or_name
Options
option_or_name may be either an object name or one of the following options:
@all | Close down everything. This is the same as clicking on from the EViews main menu. |
@objects | Close down all objects. This is the same as clicking on from the EViews main menu. |
@wf | Close down all open workfiles. |
@db | Close down all open databases. |
@prg | Close down all open program files. |
Examples
close gdp graph1 table2
closes the three objects GDP, GRAPH1, and TABLE2.
lwage.hist
close lwage
opens the LWAGE window and displays the histogram view of LWAGE, then closes the window.
close @all
closes all windows within EViews.
close @objects
closes all objects in EViews, leaving workfiles, programs, and database windows open.
Cross-references
See
“Introduction” for a discussion of basic control of EViews.
Perform either (1) Johansen’s system cointegration test, (2) Engle-Granger or Phillips-Ouliaris single equation cointegration testing, or (3) Pedroni, Kao, or Fisher panel cointegration testing for the specified series.
Syntax
There are three forms for the coint command which depend on the form of the test you wish to perform:
Johansen Cointegration Test Syntax
coint(test_option, n, option) ser1 ser2 [...ser3 ser4 ...] [@ x1 x2 x3 ...]
uses the coint keyword followed by the test_option and the number of lags n, and if desired, an “@”-sign followed by a list of exogenous variables. The first option must be one of the following six test options:
a | No deterministic trend in the data, and no intercept or trend in the cointegrating equation. |
b | No deterministic trend in the data, and an intercept but no trend in the cointegrating equation. |
c | Linear trend in the data, and an intercept but no trend in the cointegrating equation. |
d | Linear trend in the data, and both an intercept and a trend in the cointegrating equation. |
e | Quadratic trend in the data, and both an intercept and a trend in the cointegrating equation. |
s | Summarize the results of all 5 options (a-e). |
Options for the Johansen Test
restrict | Impose restrictions as specified by the append (coint) proc. |
m = integer | Maximum number of iterations for restricted estimation (only valid if you choose the restrict option). |
c = scalar | Convergence criterion for restricted estimation. (only valid if you choose the restrict option). |
save = mat_name | Stores test statistics as a named matrix object. The save= option stores a 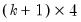 matrix, where 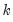 is the number of endogenous variables in the VAR. The first column contains the eigenvalues, the second column contains the maximum eigenvalue statistics, the third column contains the trace statistics, and the fourth column contains the log likelihood values. The i-th row of columns 2 and 3 are the test statistics for rank 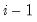 . The last row is filled with NAs, except the last column which contains the log likelihood value of the unrestricted (full rank) model. |
cvtype=ol | Display 0.05 and 0.01 critical values from Osterwald-Lenum (1992). This option reproduces the output from version 4. The default is to display critical values based on the response surface coefficients from MacKinnon-Haug-Michelis (1999). Note that the argument on the right side of the equals sign are letters, not numbers 0-1). |
cvsize=arg (default=0.05) | Specify the size of MacKinnon-Haug-Michelis (1999) critical values to be displayed. The size must be between 0.0001 and 0.9999; values outside this range will be reset to the default value of 0.05. This option is ignored if you set “cvtype=ol”. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
This type of cointegration testing may be used in a non-panel workfile. For Fisher combined testing using the Johansen framework, see below. The remaining options for the Johansen cointegration test are outlined below (
“Options for the Johansen Test”).
Note that the output for cointegration tests displays p-values for the rank test statistics. These p-values are computed using the response surface coefficients as estimated in MacKinnon, Haug, and Michelis (1999). The 0.05 critical values are also based on the response surface coefficients from MacKinnon-Haug-Michelis. Note: the reported critical values assume no exogenous variables other than an intercept and trend.
Single Equation Test Syntax
coint(method=arg, options) ser1 ser2 [...ser3 ser4 ...] [@determ determ_spec] [@regdeterm regdeterm_spec]
where
method=arg | Test method: Engle-Granger residual test (“eg”), Phillips-Ouliaris residual test (“po”). |
Cointegrating equation specifications that include a constant, linear, or quadratic trends, should use the “trend=” option to specify those terms. If any of those terms are in the stochastic regressors equations but not in the cointegrating equation, they should be specified using the “regtrend=” option.
Deterministic trend regressors that are not covered by the list above may be specified using the keywords @determ and @regdeterm. To specify deterministic trend regressors that enter into the regressor and cointegrating equations, you should add the keyword @determ followed by the list of trend regressors. To specify deterministic trends that enter in the regressor equations but not the cointegrating equation, you should include the keyword @regdeterm followed by the list of trend regressors.
Note that the p-values for the test statistics are based on simulations, and do not account for any user-specified deterministic regressors.
This type of cointegration testing may be used in a non-panel workfile. The remaining options for the single equation cointegration tests are outlined below.
Options for Single Equation Tests
Options for the Engle-Granger Test
The following options determine the specification of the Engle-Granger test (Augmented Dickey-Fuller) equation and the calculation of the variances used in the test statistic.
trend=arg (default=“const”) | Specification for the powers of trend to include in the cointegrating equation: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regtrend=arg (default=“none”) | Additional trends to include in the regressor equations (but not the cointegrating equation): None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Only trend orders higher than those specified by “trend=” will be considered. Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
lag=arg (default=“a”) | Method of selecting the lag length (number of first difference terms) to be included in the regression: “a” (automatic information criterion based selection), or integer (user-specified lag length). |
lagtype=arg (default=“sic”) | Information criterion or method to use when computing automatic lag length selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “msaic” (Modified Akaike), “msic” (Modified Schwarz), “mhqc” (Modified Hannan-Quinn), “tstat” (t-statistic). |
maxlag=integer | Maximum lag length to consider when performing automatic lag-length selection default= 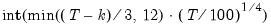 where 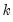 is the number of coefficients in the cointegrating equation. Applicable when “lag=a”. |
lagpval=number (default=0.10) | Probability threshold to use when performing automatic lag-length selection using a t-test criterion. Applicable when both “lag=a” and “lagtype=tstat”. |
nodf | Do not degree-of-freedom correct estimates of the variances. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Options for the Phillips-Ouliaris Test
The following options control the computation of the symmetric and one-sided long-run variances in the Phillips-Ouliaris test.
Basic Options:
trend=arg (default=“const”) | Specification for the powers of trend to include in the cointegrating equation: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regtrend=arg (default=“none”) | Additional trends to include in the regressor equations (but not the cointegrating equation): None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Only trend orders higher than those specified by “trend=” will be considered. Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
HAC Whitening Options:
lag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Kernel Options:
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
bw=arg (default=“nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwint | Use integer portion of bandwidth. |
Panel Syntax
coint(option) ser1 ser2 [...ser3 ser4 ...]
The coint command tests for cointegration among the series in the group. The second form of the command should be used with panel structured workfiles.
Options for the Panel Tests
For panel cointegration tests, you may specify the type using one of the following keywords:
Pedroni (default) | Pedroni (1994 and 2004). |
Kao | Kao (1999) |
Fisher | Fisher - pooled Johansen |
Depending on the type selected above, the following may be used to indicate deterministic trends:
const (default) | Include a constant in the test equation. Applicable to Pedroni and Kao tests. |
trend | Include a constant and a linear time trend in the test equation. Applicable to Pedroni tests. |
none | Do not include a constant or time trend. Applicable to Pedroni tests. |
a, b, c, d, or e | Indicate deterministic trends using the “a”, “b”, “c”, “d”, and “e” keywords, as detailed above in
“Options for the Johansen Test”. Applicable to Fisher tests. |
Additional Options:
ac=arg (default=“bt”) | Method of estimating the frequency zero spectrum: “bt” (Bartlett kernel), “pr” (Parzen kernel), “qs” (Quadratic Spectral kernel). Applicable to Pedroni and Kao tests. |
band=arg (default=“nw”) | Method of selecting the bandwidth, where arg may be “nw” (Newey-West automatic variable bandwidth selection), or a number indicating a user-specified common bandwidth. Applicable to Pedroni and Kao tests. |
lag=arg | For Pedroni and Kao tests, the method of selecting lag length (number of first difference terms) to be included in the residual regression. For Fisher tests, a pair of numbers indicating lag. |
infosel=arg (default=“sic”) | Information criterion to use when computing automatic lag length selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn). Applicable to Pedroni and Kao tests. |
maxlag=int | Maximum lag length to consider when performing automatic lag length selection, where int is an integer. The default is where 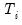 is the length of the cross-section. Applicable to Pedroni and Kao tests. |
disp=arg (default=500) | Maximum number of individual results to be displayed. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
Johansen test
coint(s,4) x y z
summarizes the results of the Johansen cointegration test for the series X, Y, and Z for all five specifications of trend. The test equation uses lags of up to order four.
Engle-Granger Test
coint(method=eg) x y z
performs the default Engle-Granger test on the residuals from a cointegrating equation which includes a constant. The number of lags is determined using the SIC criterion and an observation-based maximum number of lags.
coint(method=eg, trend=linear, lag=a, lagtype=tstat, lagpval=.15, maxlag=10) x y z
employs a cointegrating equation that includes a constant and linear trend, and uses a sequential t-test starting at lag 10 with threshold probability 0.15 to determine the number of lags.
coint(method=eg, lag=5) x y z
conducts an Engle-Granger cointegration test on the residuals from a cointegrating equation with a constant, using a fixed lag of 5.
Phillips-Ouliaris Test
coint(method=po) x y z
performs the default Phillips-Ouliaris test on the residuals from a cointegrating equation with a constant, using a Bartlett kernel and Newey-West fixed bandwidth.
coint(method=po, bw=andrews, kernel=quadspec, nodf) x y z
estimates the long-run covariances using a Quadratic Spectral kernel, Andrews automatic bandwidth, and no degrees-of-freedom correction.
coint(method=po, trend=linear, lag=1, bw=4) x y z
estimates a cointegrating equation with a constant and linear trend, and performs the Phillips-Ouliaris test on the residuals by computing the long-run covariances using AR(1) prewhitening, a fixed bandwidth of 4, and the Bartlett kernel.
Panel Tests
For a panel structured workfile,
coint(pedroni,maxlag=3,infosel=sic) x y z
performs Pedroni’s residual-based panel cointegration test with automatic lag selection with a maximum lag limit of 3. Automatic selection based on Schwarz criterion.
Cross-references
See
“Cointegration Testing” for details on the various cointegration tests.
See
Equation::coint and
Group::coint for the related object routines.
Estimate a cointegrating equation using Fully Modified OLS (FMOLS), Canonical Cointegrating Regression (CCR), or Dynamic OLS (DOLS) in single time series settings, and Panel FMOLS and DOLS in panel workfiles.
Syntax
cointreg(options) y x1 [x2 x3 ...] [@determ determ_spec] [@regdeterm regdeterm_spec]
List the coint keyword, followed by the dependent variable and a list of the cointegrating variables.
Cointegrating equation specifications that include a constant, linear, or quadratic trends, should use the “trend=” option to specify those terms. If any of those terms are in the stochastic regressors equations but not in the cointegrating equation, they should be specified using the “regtrend=” option.
Deterministic trend regressors that are not covered by the list above may be specified using the keywords @determ and @regdeterm. To specify deterministic trend regressors that enter into the regressor and cointegrating equations, you should add the keyword @determ followed by the list of trend regressors. To specify deterministic trends that enter in the regressor equations but not the cointegrating equation, you should include the keyword @regdeterm followed by the list of trend regressors.
Basic Options
method=arg (default=“fmols”) | Estimation method: Fully Modified OLS (“fmols”), Canonical Cointegrating Regression (“ccr”), Dynamic OLS (“dols”) Note that CCR estimation is not available in panel settings. |
trend=arg (default=“const”) | Specification for the powers of trend to include in the cointegrating and regressor equations: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regtrend=arg (default=“none”) | Additional trends to include in the regressor equations (but not the cointegrating equation): None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Only trend orders higher than those specified by “trend=” will be considered. Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regdiff | Estimate the regressor equation innovations directly using the difference specifications. |
coef=arg | Specify the name of the coefficient vector; the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
In addition to these options, there are specialized options for each estimation method.
Panel Options
panmethod=arg (default=“pooled”) | Panel estimation method: pooled (“pooled”), pooled weighted (“weighted”), grouped (“grouped”) |
pancov=sandwich | Estimate the coefficient covariance using a sandwich method that allows for cross-section heterogeneity. |
Options for FMOLS and CCR
To estimate FMOLS or CCR use the “method=fmols” or “method=ccr” options. The following options control the computation of the symmetric and one-sided long-run covariance matrices and the estimate of the coefficient covariance.
HAC Whitening Options
lag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Kernel Options
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
bw=arg (default=“nwfixed”) | Bandwidth:: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
bwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Coefficient Covariance
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
Panel Options
hetfirst | Estimate the first-stage regression assuming heterogeneous coefficients. For FMOLS specifications estimated using pooled or pooled weighted methods (“panmethod =pooled”, “panmethod=weighted”) |
Options for DOLS
To estimate using DOLS use the “method=dols” option. The following options control the specification of the lags and leads and the estimate of the coefficient covariance.
lltype=arg (default=“fixed”) | Lag-lead method: fixed values (“fixed”), automatic selection - Akaike (“aic”), automatic - Schwarz (“sic”), automatic - Hannan-Quinn (“hqc”), None (“none”). |
lag=arg | Lag specification: integer (required user-specified number of lags if “lltype=fixed”). |
lead=arg | Lead specification: integer (required user-specified number of lags if “lltype=fixed”). |
maxll=integer | Maximum lag and lead-length for automatic selection (optional user-specified integer if “lltype=” is used to specify automatic selection). The default is an observation-based maximum. |
cov=arg | Coefficient covariance method: (default) long-run variance scaled OLS, unscaled OLS (“ols”), White (“white”), Newey-West (“hac”). |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
For the default covariance calculation or “cov=hac”, the following options control the computation of the long-run variance or robust covariance:
HAC Covariance Whitening Options (if default covariance or “cov=hac”)
covlag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Covariance Kernel Options (if default covariance or “cov=hac”)
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “covbw=neweywest”). |
covbwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
covbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Panel Options
Weighted coefficient or coefficient covariance estimation in panel DOLS requires individual estimates of the long-run variances of the residuals. You may compute these estimates using the standard default long-run variance options, or you may choose to estimate it using the ordinary variance.
For weighted estimation we have:
panwgtlag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
panwgtinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lrvarlag=a”). |
panwgtmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lrvarlag=a”). The default is an observation-based maximum. |
panwgtkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
panwgtbw=arg (default=“nwfixed”) | Bandwidth:: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
panwgtnwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
panwgtbwoffset=integer (default=0) | Apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
panwgtbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
For the coefficient covariance estimation we have:
lrvar=ordinary | Compute DOLS estimates of the long-run residual variance used in covariance calculation using the ordinary variance. |
lrvarlag=arg (default=0) | For DOLS estimates of the long-run residual variance used in covariance calculation, lag specification: integer (user-specified lag value), “a” (automatic selection). |
lrvarinfosel=arg (default=“aic”) | For DOLS estimates of the long-run residual variance used in covariance calculation, information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lrvarlag=a”). |
lrvarmaxlag=integer | For DOLS estimates of the long-run residual variance used in covariance calculation, maximum lag-length for automatic selection (optional) (if “lrvarlag=a”). The default is an observation-based maximum. |
lrvarkern=arg (default=“bart”) | For DOLS estimates of the long-run residual variance used in covariance calculation, Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
lrvarbw=arg (default=“nwfixed”) | For DOLS estimates of the long-run residual variance used in covariance calculation, bandwidth:: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
lrvarnwlag=integer | For DOLS estimates of the long-run residual variance used in covariance calculation, Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
lrvarbwoffset=integer (default=0) | For DOLS estimates of the long-run residual variance used in covariance calculation, apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
lrvarbwint | For DOLS estimates of the long-run residual variance used in covariance calculation, use integer portion of bandwidth. |
Examples
FMOLS and CCR
To estimate, by FMOLS, the cointegrating equation for LC and LY including a constant, you may use:
cointreg(nodf, bw=andrews) lc ly
The long-run covariances are estimated nonparametrically using a Bartlett kernel and a bandwidth determined by the Andrews automatic selection method. The coefficient covariances are estimated with no degree-of-freedom correction.
To include a linear trend term in a model where the long-run covariances computed using the Quadratic Spectral kernel and a fixed bandwidth of 10, enter:
cointreg(trend=linear, nodf, bw=10, kern=quadspec) lc ly
A model with a cubic trend may be estimated using:
cointreg(trend=linear, lags=2, bw=neweywest, nwlag=10, kernel=parzen) lc ly @determ @trend^3
Here, the long-run covariances are estimated using a VAR(2) prewhitened Parzen kernel with Newey-West nonparametric bandwidth determined using 10 lags in the autocovariance calculations.
cointreg(trend=quadratic, bw=andrews, lags=a, infosel=aic, kernel=none, regdiff) lc ly @regdeterm @trend^3
estimates a restricted model with a cubic trend term that does not appear in the cointegrating equation using a parametric VARHAC with automatic lag length selection based on the AIC. The residuals for the regressors equations are obtained by estimating the difference specification.
To estimate by CCR, we provide the “method=ccr” option. The command
cointreg(method=ccr, lag=2, bw=andrews, kern=quadspec) lc ly
estimates, by CCR, the constant only model using a VAR(2) prewhitened Quadratic Spectral and Andrews automatic bandwidth selection.
cointreg(method=ccr, trend=linear, lag=a, maxlag=5, bw=andrews, kern=quadspec) lc ly
modifies the previous estimates by adding a linear trend term to the cointegrating and regressors equations, and changing the VAR prewhitening to automatic selection using the default SIC with a maximum lag length of 5.
cointreg(method=ccr, trend=linear, regtrend=quadratic, lag=a, maxlag=5, bw=andrews) lc ly
adds a quadratic trend term to the regressors equations only, and changes the kernel to the default Bartlett.
DOLS
cointreg(method=dols, trend=linear, nodf, lag=4, lead=4) lc ly
estimates the linear specification using DOLS with four lags and leads. The coefficient covariance is obtained by rescaling the no d.f.-correction OLS covariance using the long-run variance of the residuals computed using the default Bartlett kernel and default fixed Newey-West bandwidth.
cointreg(method=dols, trend=quadratic, nodf, lag=4, lead=2, covkern=bohman, covbw=10) lc ly @determ @trend^3
estimates a cubic specification using 4 lags and 2 leads, where the coefficient covariance uses a Bohman kernel and fixed bandwidth of 10.
cointreg(method=dols, trend=quadratic, nodf, lag=4, lead=2, cov=hac, covkern=bohman, covbw=10) lc ly @determ @trend^3
estimates the same specification using a HAC covariance in place of the scaled OLS covariance.
cointreg(method=dols, trend=quadratic, lltype=none, cov=ols) lc ly @determ @trend^3
computes the Static OLS estimates with the usual OLS d.f. corrected coefficient covariance.
Cross-references
See
“Cointegrating Regression” for a discussion of single equation cointegrating regression. See
“Panel Cointegration Estimation” for discussion of estimation in panel settings.
See
“Technical Discussion” for a discussion of VEC estimation.
Place vector in column of a matrix.
Place a column or rowvector object in a specified column of a matrix.
Syntax
colplace(m, r, n)
Places the column vector or rowvector v into the matrix m at column n. The number of rows of m and v must match, and the destination column must already exist within m.
Examples
matrix m1 = @mnrnd(30, 5)
vector v1 = @mnrnd(30)
colplace(m1, v1, 3)
The third column of M1 will be set equal to the vector V1.
Cross-references
Add comment to command capture window.
Syntax
commandcap arg
sends arg to the command capture window. Of particular use to add-in writers for sending notifications to users.
Cross-references
See
“Command Capture” for further details.
Copy an object, or a set of objects matching a name pattern, within and between workfiles, workfile pages, and databases. Data in series objects may be frequency converted or match merged.
Syntax
copy(options) src_spec dest_spec [src_id dest_id]
copy(options) src_spec dest_spec [@src src_ids @dest dest_id]
where src_spec and dest_spec are of the form:
[ctype][container::][page\]object_name
There are three parts to the copy command: (1) a specification of the location and names of the source objects; (2) a specification of the location and names of the destination objects; (3) optional source and destination IDs if the copy operation involves match merging.
The source and destination objects are specified in multiple (optional) parts: (1) the container specification is the name of a workfile or database; (2) the page specification is the name of a page within a workfile or a subdirectory within a database; and (3) the object_name specification is the name of an object or a wildcard pattern corresponding to multiple objects.
The ctype specification is rarely required, but permits you to specify precisely your source or destination in cases where a database and workfile share the same name. In this case, ctype may be used to indicate the container to which you are referring by prefixing the container name with “:” to indicate the workfile, or “::” to indicate the database with the common name.
When parts of the source or destination specification are not provided, EViews will fill in default values where possible. The default container is the active workfile, unless the “::” prefix is used in which case the default container is the default database. The default page within a workfile is always the active page. The default name for the destination object is the name of the object within the source container.
If ID series are not provided in the command, then EViews will perform frequency conversion when copying data whenever the source and destination containers have different frequencies. If ID series are provided, then EViews will perform a general match merge between the source and destination using the specified ID series. In the case where you wish to copy your data using match merging with special treatment for date matching, you must use the special keyword “@DATE” as an ID series for the source or destination. If “@DATE” is not specified as an identifier in either the source or destination IDs, EViews will perform an exact match merge using the given identifiers.
If ID series are not specified, but a conversion option requiring a general match merge is used (e.g., “c=med”), “@DATE @DATE” will be appended to the list of IDs and a general date match merge will be employed.
See
Link::linkto for additional discussion of the differences embodied in these choices.
The general syntax described above covers all possible uses of the copy command. The following paragraphs provide examples of the specific syntax used for some common cases of the command.
Copying Within a Workfile
Copy an object within the default workfile page as a new object with a different name:
• copy(options) src_name dest_name
Copy an object from the src_page page into the default workfile page using the specified name:
• copy(options) src_page\src_name dest_name
Copy an object from the src_page page into the dest_page page, keeping the same name:
• copy(options) src_page\src_names dest_page\
Copy an object from the src_page page to the default workfile page, match merging any series data using a single src_id and a single dest_id identifier series:
• copy(options) src_page\src_name dest_name src_id dest_id
Copy an object from the src_page page to the dest_page match merging any series data using multiple source and destination identifier series:
• copy(options) src_page\src_name dest_page\dest_name @src src_id1 src_id2 ... src_id_n @dest dest_id1 dest_id2 ... dest_id_n
Copying Between Containers (Workfiles and Databases)
Copy one or more objects from the src_page of the workfile src_workfile to the dest_page of the workfile dest_workfile, using the name or name pattern given in src_names:
• copy(options) src_workfile::src_page\src_names dest_workfile::dest_page\
Copy an object from database src_database to the default page in the container dest_container:
• copy(options) src_database::src_name dest_container::dest_name
Note that if both a workfile and database exist matching the name provided in dest_container, EViews will favor the workfile unless the “::” prefix is used to specify explicitly that the database should be used.
Options
Basic Options
overwrite / o | Overwrite any existing object with the destination name in the destination container. Error only if a non-editable series is encountered in the destination location. |
merge / m | If the source object is a series, merge the data from the source series into any existing destination series, preserving any values in the destination series that are not present in the source. For all other object types, overwrite any existing object with the source object. Error if a non-editable series is encountered in the destination location. |
protect / p | Protect objects in the destination location from overwriting or merging. If there is an existing object in the destination container, cancel the copy operation for that object, but do not generate an error. |
noerr | Suppress errors that are generated during the copy. For example, if the overwrite option is used, suppress any error caused by attempting to overwrite a non-editable series such as an index series used in the workfile structure. |
link | Link the object to the source data so that the values can be refreshed at a later time. |
Group Copy Options
When copying a group object from workfile to database:
g=arg | Method for copying group objects from a workfile to database: “s” (copy group definition and series as separate objects), “t” (copy group definition and series as one object), “d” (copy series only as separate objects), “l” (copy group definition only). |
When copying a group object from a database to a workfile:
g=arg | Method for copying group objects from a database or workfile to a workfile: “b” (copy both group definition and series), “d” (copy only the series), “l” (copy only the group definition). |
Note that copying a group object containing expressions or auto-updating series between workfiles only copies the expressions, and not the underlying series.
Frequency Conversion Options
If the copy command does not specify identifier series, EViews will perform frequency conversion of the data contained in series objects whenever the source and destination containers are dated, but do not have the same frequency.
If either of the pages are undated, EViews will, unless match merge options are provided (as described below), perform a raw copy, in which the first observation in the source workfile page is copied into the first observation in the destination page, the second observation in the source into the second observation in the destination, and so forth.
The following options control the frequency conversion method when copying series and group objects into a workfile page and converting from low to high frequency:
c=arg | Low to high conversion methods: “r” or “repeata” (constant match average), “d” or “repeats” (constant match sum), “q” or “quada” (quadratic match average), “t” or “quads” (quadratic match sum), “linearf” (linear match first), “i” or “linearl” (linear match last), “cubicf” (cubic match first), “c” or “cubicl” (cubic match last), “pointf” (point first), “pointl” (point last), “dentonf” (Denton first), “dentonl” (Denton last), “dentona” (Denton average), “dentons” (Denton sum), “chowlinf” (Chow-Lin first), “chowlinl” (Chow-Lin last), “chowlina” (Chow-Lin average),“chowlins” (Chow-Lin sum), “litmanf” (Litterman first), “litmanl” (Litterman last), “litmana” (Litterman average), “litmans” (Litterman sum). |
rho=arg | Autocorrelation coefficient (for Chow-Lin and Litterman conversions). Must be between 0 and 1, inclusive. |
In addition, for Denton, Chow-Lin, and Litterman conversions, you must specify the indicator series by appending the keyword “@indicator” followed by the series name at the end of the copy command.
The following options control the frequency conversion method when copying series and group objects into a workfile page and converting from high to low frequency:
c=arg | High to low conversion methods removing NAs: “a” (average of the nonmissing observations), “s” (sum of the nonmissing observations), “f” (first nonmissing observation), “l” (last nonmissing observation), “x” (maximum nonmissing observation), “m” (minimum nonmissing observation). High to low conversion methods propagating NAs: “an” or “na” (average, propagating missings), “sn” or “ns” (sum, propagating missings), “fn” or “nf” (first, propagating missings), “ln” or “nl” (last, propagating missings), “xn” or “nx” (maximum, propagating missings), “mn” or “nm” (minimum, propagating missings). |
Note that if no conversion method is given in the command, the conversion method specified within the series object will be used as the default. If the series does not contain an explicit conversion method, the global option settings will used to determine the method.
Frequency conversion involving panel structured pages involves special handling:
• If both pages are dated panel pages that are structured with a single identifier, EViews will perform frequency conversion cross-section by cross-section.
• Conversion from a dated panel page to a dated, non-panel page will first perform a mean contraction across cross-sections to obtain a single time series (by computing the means for each period), and then a frequency conversion of the resulting time series to the new frequency.
• Conversion from a dated, non-panel page to a dated panel page will first involve a frequency conversion of the single time series to the new frequency. The converted time series will be used for each cross-section in the panel page.
In all three of these cases, all of the high-to-low conversion methods are supported, but low-to-high frequency conversion only offers (repeating of the low frequency observations).
Lastly, conversion involving a panel page with more than one dimension or an undated page will default to raw data copy unless general match merge options are provided.
Match Merge Options
These options are available when ID series are specified in the copy commmand.
smpl= smpl_spec | Sample to be used when computing contractions during copying using match merge. Either provide the sample range in double quotes or specify a named sample object. By default, EViews will use the entire workfile sample “@ALL”. |
c=arg | Set the match merge contraction method. If you are copying a numeric source series by general match merge, the argument can be one of: “mean”, “med” (median), “max”, “min”, “sum”, “sumsq” (sum-of-squares), “var” (variance), “sd” (standard deviation), “skew” (skewness), “kurt” (kurtosis), “quant” (quantile, used with “quant=” option), “obs” (number of observations), “nas” (number of NA values), “first” (first observation in group), “last” (last observation in group), “unique” (single unique group value, if present), “none” (disallow contractions). If copying an alpha series, only the non-summary methods “max”, “min”, “obs”, “nas”, first”, “last”, “unique” and “none” are supported. For copying of numeric series, the default contraction method is “c=mean”; for copying of alpha series, the default is “c=unique”. |
quant=number | Quantile value to be used when contracting using the “c=quant” option (e.g, “quant=.3”). |
nacat | Treat “NA” values as a category when copying using general match merge operations. |
Most of the conversion options should be self-explanatory. As for the others: “first” and “last” give the first and last non-missing observed for a given group ID; “obs” provides the number of non-missing values for a given group; “nas” reports the number of NAs in the group; “unique” will provide the value in the source series if it is the identical for all observations in the group, and will return NA otherwise; “none” will cause the copy to fail if there are multiple observations in any group—this setting may be used if you wish to prohibit all contractions.
On a match merge expansion, copying with match merging will repeat the value of the source for every observation with matching identifier values in the destination. If both the source and destination have multiple values for a given ID, EViews will first perform a contraction across IDs in the source (if not ruled out by “c=none”), and then perform the expansion by replicating the contracted value in the destination. For example, converting from a quarterly panel to an annual panel using match merge, EViews will first contract the data across IDs down to a single quarterly time series, will convert the series to an annual frequency, then will assign the annual data to each of the cross-sections in the destination page.
Examples
copy good_equation best_equation
makes an exact copy of GOOD_EQUATION and names it BEST_EQUATION.
copy graph_1 wf2::wkly\graph1
copies GRAPH_1 from the default page of the current workfile to GRAPH1 in the page WKLY of the workfile WF2.
copy gdp usdat::
copies GDP from the current workfile to GDP in the USDAT database or workfile.
copy ::gdp macro1::gdp_us
copies GDP from the default database to either the open workfile MACRO1, or the database named MACRO1 if there is no open workfile with that name. If there is an open workfile MACRO1 you may use
copy ::gdp ::macro1::gdp_us
to specify explicitly that you wish to write to the MACRO1 database.
copy(smpl="1990 2000") page1\pop page2\ @src county @date @dest county @date
copies POP data for 1990 through 2005 from PAGE1 to PAGE2, match merge using the ids COUNTY and the date structure of the two pages.
copy(smpl="1990 2000", c=mean) panelpage\inc countypage\ county county
copies the INC data from the PANELPAGE to the COUNTYPAGE, match merging using the values of the COUNTY series, and contracting the panel data by computing means for each county using the specified sample.
copy countypage\pop panelpage\ county county
match merges the POP data from the COUNTYPAGE to the PANELPAGE using the values of the COUNTY series.
copy(c=x, merge) quarterly::page1\ser* annual::page6\*
copies all objects with names beginning with “SER” on page PAGE1 of workfile QUARTERLY into page PAGE6 of workfile ANNUAL using the existing names. Series objects with data that can be (high-to-low) frequency converted will take the maximum value within a low-frequency period as the conversion method. If destination series already exist with the same name as the source series, the data will be merged. If destination objects (non-series) exist with the same name as source series, they will be overwritten.
Note that since databases are read from disk, you may provide a path for the database in the container specification, as in:
copy "c:\my data\dba.edb::ser01" ser02
which copies the object SER01 from the database DBA.EDB located in the path “C:\MY DATA\” to SER02 in the default workfile page.
copy gd* "c:\my data\findat::"
makes a duplicate of all objects in the default page of the current workfile with names starting with “GD” to the database FINDAT in the root of “C:\MY DATA\”.
Cross-references
See
“Copying Objects” for a discussion of copying and moving objects.
Compute Pearson product-moment (ordinary) correlations for the specified series or groups.
Syntax
cor(options) arg1 [arg2 arg3...]
where arg1, arg2, etc. are the names of series or groups.
Note that this command is a limited feature version of the group view
Group::cor.
Options
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
out=name | Basename for saving output. All results will be saved in Sym matrices named using the string “CORR”, appended to the basename (e.g., the correlation specified by “out=my” is saved in the Sym matrix “MYCORR”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
cor height weight age
displays a
Pearson correlation matrix for the three series HEIGHT, WEIGHT, and AGE.
Cross-references
See
Group::cor for more general routines for computing correlations.
See also
cov. For simple functions to perform the calculations, see
@cor, and
@cov.
Estimates models where the dependent variable is a nonnegative integer count.
Syntax
count(options) y x1 [x2 x3...]
count(options) specification
Follow the count keyword by the name of the dependent variable and a list of regressors.
Options
d=arg (default=“p”) | Likelihood specification: Poisson likelihood (“p”), normal quasi-likelihood (“n”), exponential likelihood (“e”), negative binomial likelihood or quasi-likelihood (“b”). |
v=positive_num (default=1) | Specify fixed value for QML parameter in normal and negative binomial quasi-likelihoods. |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich methods)., “glm” (GLM method).. |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
g | GLM standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of the EViews default values (out of range values are set to “s=1”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
The command:
count(d=n,v=2,cov=glm) y c x1 x2
estimates a normal QML count model of Y on a constant, X1, and X2, with fixed variance parameter 2, and GLM standard errors.
count arrest c job police
makeresid(g) res_g
estimates a Poisson count model of ARREST on a constant, JOB, and POLICE, and stores the generalized residuals in the series RES_G.
count(d=p) y c x1
fit yhat
estimates a Poisson count model of Y on a constant and X1, and saves the fitted values (conditional mean) in the series YHAT.
count(d=p, h) y c x1
estimates the same model with QML standard errors and covariances.
Cross-references
See
“Count Models” for additional discussion.
See
Equation::count for the equivalent equation object command.
Compute Pearson product-moment (ordinary) covariances for the specified series or groups.
Syntax
cor arg1 [arg2 arg3...]
where arg1, arg2, etc. are the names of series or groups.
Note that this command is a limited feature version of the group view
Group::cov.
Options
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
out=name | Basename for saving output. All results will be saved in Sym matrices named using the string “CORR”, appended to the basename (e.g., the correlation specified by “out=my” is saved in the sym matrix “MYCORR”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
cov height weight age
displays a
Pearson covariance matrix for the three series HEIGHT, WEIGHT, and AGE.
Cross-references
See
Group::cov for more general routines for computing covariances.
See also
cor .For simple functions to perform the calculations, see
@cor, and
@cov.
Create workfile.
This command has been replaced by
wfcreate and
pagecreate.
Displays cross correlations (correlograms) for a pair of series.
Syntax
cross(n,options) ser1 ser2 [ser3 ...]
You must specify the number of lags n to use in computing the cross correlations as the first option. EViews will create an untitled group from the specified series and groups, and will display the cross correlation view for the group.
Options
The following options may be specified inside the parentheses after the number of lags:
prompt | Force the dialog to appear from within a program. |
p | Print the cross correlogram. |
Examples
cross(36) log(m1) dlog(cpi)
displays the cross correlogram between the log of M1 and the first difference of the log of CPI, using up to 36 leads and lags.
equation eq1.arch sp500 c
eq1.makeresid(s) res_std
cross(24) res_std^2 res_std
The first line estimates a GARCH(1,1) model and the second line retrieves the standardized residuals. The third line plots the cross correlogram squared standardized residual and the standardized residual, up to 24 leads and lags. This correlogram provides a rough check of asymmetry in the ARCH effect.
Cross-references
See
Group::cross for the equivalent group view command.
Enter data from keyboard.
Opens an unnamed group window to edit one or more series.
Syntax
data arg1 [arg2 arg3 ...]
Follow the data keyword by a list of series and group names. You can list existing names or new names. Unrecognized names will cause new series to be added to the workfile. These series will be initialized with the value “NA”.
Examples
data group1 newx newy
opens a group window containing the series in group GROUP1, and the series NEWX and NEWY.
Cross-references
See
“Entering Data” for a discussion of the process of entering data from the keyboard.
Open or create a database.
If the specified database does not exist, a new (empty) database will be created and opened. The opened database will become the default database.
Syntax
db(options) [path\]db_name [as shorthand_name]
Follow the db command by the name of the database to be opened or to be created (if it does not already exist). You may include a path name to work with a database not in the default path.
You may use the “as” keyword to provide an optional shorthand_name or short text label which may be used to refer to the database in commands and programs. If you leave this field blank, a default shorthand_name will be assigned automatically.
See
“Database Shorthands” for additional discussion.
Options
See
dbopen for a list of available options for working with foreign format databases.
Examples
db findat
opens the database FINDAT in the default path and makes it the default database from which to store and fetch objects. If the database FINDAT does not already exist, an empty database named FINDAT will be created and opened.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
See also
dbcreate and
dbopen.
Make a copy of an existing database.
Syntax
dbcopy [path\]source_name [path\]copy_name
Follow the dbcopy command by the name of the existing database and a name for the copy. You should include a path name to copy from or to a database that is not in the default directory. All files associated with the database will be copied.
Options
type=arg | Specify the source database type: (see table below). The default is to read an EViews 7 database. |
desttype=arg, t=arg | Specify the destination database type: (see table below). The default is to create an EViews 7 database. |
The following table summaries the various database formats, along with the corresponding allowable “type=” and “desttype=” keywords:
| |
Aremos-TSD x | “a”, “aremos”, “tsd” |
DRIBase x | “b” “dribase” |
DRIPro Link x | “dripro” |
DRI DDS | “dds” |
EViews | “e”, “evdb” |
EViews 6 compatible | “eviews6” |
FAME | “f”, “fame” |
GiveWin/PcGive | “g”, “give” |
RATS 4.x | “r”, “rats” |
RATS Portable / TROLL | “l”, “trl” |
TSP Portable | “t”, “tsp” |
For the source specification, the following options may be required when connecting to a remote server:
s=server_id, server=server_id | Server name |
u=user, username=user | Username |
p=pwd, password=pwd | Password |
For the destination specification, the following options may be required when connecting to a remote server:
dests=server_id, destserver =server_id | Server name |
destu=user, destusername=user | Username |
destp=pwd, destpassword =pwd | Password |
Examples
dbcopy usdat c:\backup\usdat
makes a copy of all files associated with the database USDAT in the default path and stores it in the “c:\backup” directory under the basename “Usdat”.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
Create a new database.
Syntax
dbcreate(options) [path\]db_name [as shorthand_name]
Follow the dbcreate keyword by a name for the new database. You may include a path name to create a database not in the default directory. The new database will become the default database.
You may use the “as” keyword to provide an optional
shorthand_name or a short text label which may be used to refer to the open database in commands and programs. If you leave this field blank, a default
shorthand_name will be assigned automatically. See
“Database Shorthands” for additional discussion.
Options
type=arg, t=arg | Specify the database type: (see table below). The default is to create an EViews 7 database. |
The following table summaries the various database formats, along with the corresponding “type=” keywords:
| |
Aremos-TSD x | “a”, “aremos”, “tsd” |
DRIBase x | “b” “dribase” |
DRIPro Link x | “dripro” |
DRI DDS | “dds” |
EViews | “e”, “evdb” |
EViews 6 compatible | “eviews6” |
FAME | “f”, “fame” |
GiveWin/PcGive | “g”, “give” |
RATS 4.x | “r”, “rats” |
RATS Portable / TROLL | “l”, “trl” |
TSP Portable | “t”, “tsp” |
DRIBase and FAME databases are not supported in EViews Standard Edition.
The following options may be required when connecting to a remote server:
s=server_id, server=server_id | Server name |
u=user, username=user | Username |
p=pwd, password=pwd | Password |
Examples
dbcreate macrodat
creates a new database named MACRODAT in the default path, and makes it the default database from which to store and fetch objects. This command will issue an error message if a database named
MACRODAT already exists. To open an existing database, use
dbopen or
db.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
Delete an existing database (all files associated with the specified database).
Syntax
dbdelete [path\]db_name
Follow the dbdelete keyword by the name of the database to be deleted. You may include a path name to delete a database not in the default path.
Examples
dbdelete c:\temp\testdat
deletes all files associated with the TESTDAT database in the specified directory.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
See also
dbcopy and
dbdelete.
Open an existing database.
Syntax
dbopen(options) [path\]db_name [as shorthand_name]
Follow the dbopen keyword with the name of a database. You should include a path name to open a database not in the default path. The opened database will become the default database.
You do not need to specify a database name when opening a Datastream or FRED connection (“type=datastream” or “type=fred”) as EViews will automatically connect to the proper location.
You may use the “as” keyword to provide an optional shorthand_name or a short text label which is used to refer to the open database in commands and programs. If you leave this field blank, a default shorthand_name will be assigned automatically.
See
“Database Shorthands” for additional discussion.
By default, EViews will use the extension of the database file to determine type. For example, files with the extension “.EDB” will be opened as an EViews database. You may use the “type=” option to specify an explicit type.
Options
type=arg, t=arg | Specify the database type: (see table below). |
The following table summaries the various database formats, along with the corresponding “type=” keywords:
| | |
Australian Bureau of Statistics SDMX | “abs” | (b) |
Bloomberg | “bloom” | (a), (b) |
Bureau of Economic Analysis | “bea” | (b) |
Bureau of Labor Statistics | “bls” | (b) |
CEIC | “ceic” | (a), (b) |
Datastream | “datastream” | (a), (b) |
DBnomics | “dbnomics” | (b) |
Deutsche Bundesbank SDMX | “bbk” | (b) |
ECB (European Central Bank) | ecb” | (b) |
EIA Bulk File | “eiabulk” | (a), (c) |
EIA (U.S. Energy Information Administration) | “eia” | (a), (b) |
Eurostat SDMX | “eurostat” | (b) |
EViews | “e”, “eviews” | |
FAME | “f”, “fame” | (a) |
FRED | “fred” | (b) |
FRED v1 | “fredv1” | (b) |
Haver | “h”, “haver” | (a) |
IHS Global Insight | “ihs global insight” | (a), (b) |
IHS Magellan | “magellan” | (a), (b) |
IHSMarkit API | “ihsmarkit” | (a), (b) |
IMF (International Monetary Fund) SDMX | “imf” | (b) |
INSEE (National Institute of Statistics and Economic Studies) SDMX | “insee” | (b) |
Moody’s Economy.com | “economy” | (a), (b) |
NOAA (National Oceanic And Atmospheric Administration) | “noaa” | (b) |
OECD (Organization for Economic Cooperation and Development) SDMX | “oecd” | (b) |
SDMX_ML | “sdmx” | (a), (c) |
StatCan SDMX (Statistics Canada SDMX) | “statcan” | (a), (b) |
Trading Economics | “tradingeconomics” | (a), (b) |
TSP Portable | “t”, “tsp” | |
UN (United Nations) | “un” | (b) |
US Census | “uscensus” | (b) |
WHO (World Health Organization) | “who” | (a), (b) |
World Bank | “worldbank” | (b) |
• (a) Not supported in EViews Standard Edition.
• (b) You must have an active connection to the internet to access these databases.
• (c) You must have internet access to download these file-based databases prior to opening them with EViews.
In addition, specific types may require installation of additional software. For details see,
“Notes on Particular Formats”.
The following options may be required when connecting to a remote server:
s=server_id, server=server_id | Server name |
u=user, username=user | Username |
p=pwd, password=pwd | Password |
Examples
dbopen c:\data\us1
opens a database named US1 in the C:\DATA directory. The command:
dbopen us1
opens a database in the default path. If the specified database does not exist, EViews will issue an error message. You should use
db or
dbcreate to create a new database.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
See also
db and
dbcreate.
Pack an existing database.
Syntax
dbpack [path\]db_name
Follow the dbpack keyword by a database name. You may include a path name to pack a database not in the default path.
Examples
dbpack findat
packs the database named FINDAT in the default path.
Cross-references
See
“Packing the Database” for additional discussion.
Rebuild an existing database.
Rebuild a seriously damaged database into a new database file.
Syntax
dbrebuild [path\]source_name [path\]dest_name
Follow the dbrebuild keyword by the name of the database to be rebuilt, and then a new database name.
Examples
If you issue the command:
dbrebuild testdat fixed_testdat
EViews will attempt to rebuild the database TESTDAT into the database FIXED_TESTDAT in the default directory.
Cross-references
Rename an existing database.
dbrename renames all files associated with the specified database.
Syntax
dbrename [path\]old_name [path\]new_name
Follow the dbrename keyword with the current name of an existing database and the new name for the database.
Examples
dbrename testdat mydat
Renames all files associated with the TESTDAT database in the specified directory to MYDAT in the default directory.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
See
db and
dbcreate. See also
dbcopy and
dbdelete.
Deletes objects from a workfile or a database.
Syntax
delete(options) arg1 [arg2 arg3 ...]
Follow the keyword by a list of the names of any objects you wish to remove from the current workfile. Deleting does not remove objects that have been stored on disk in EViews database files.
Options
noerr | Do not error if the object doesn’t exist. |
You can delete an object from a database by prefixing the name with the database name and a double colon. You can use a pattern to delete all objects from a workfile or database with names that match the pattern. Use the “?” to match any one character and the “*” to match zero or more characters.
If you use delete in a program file, EViews will delete the listed objects without prompting you to confirm each deletion.
Examples
To delete all objects in the workfile with names beginning with “TEMP”, you may use the command:
delete temp*
To delete the objects CONS and INVEST from the database MACRO1, use:
delete macro1::cons macro1::invest
Cross-references
See
“Object Commands” for a discussion of working with objects.
See
“EViews Databases” for a discussion of EViews databases.
Unregister a program file as an EViews Add-in.
Syntax
deleteaddin(options) [path\]prog_name
unregisters the specified program file as an EViews Add-in.
If you do not provide the optional path specification, EViews looks for the program file in the default EViews Add-ins directory.
Explicit path specifications containing “.\” and “..\” (to indicate the current level and one directory level up) are evaluated relative EViews default directory.
You may use the special “<addins>” directory keyword in your path specification.
Options
type=arg | Specify the Add-ins type, where arg is the name of a EViews object type. The default is to create a global Add-in. |
proc=arg | User-defined command/procedure name. If omitted, the Add-in will not have a command form. |
Examples
deleteaddin .\myaddin.prg
unregisters the Add-in associated with file “Myaddin.prg”.
Alternatively,
deleteaddin(proc="myaddin")
unregisters the Add-in whose proc name matches “myaddin”. Note that this name may not match the program name.
deleteaddin(type="graph", proc="recshade")
unregisters the graph “Recshade” specific Add-in. In cases, where more than 1 Add-in has the same proc name, the type is useful to differentiate which is to be unregistered.
Cross-references
See
“Add-ins” for a detailed discussion of Add-ins.
Estimate a equation in a panel structured workfile using the difference-in-difference estimator.
Syntax
did(options) y [x1] [@ treatment]
List the dependent variable, followed by an optional list of exogenous regressors, followed by an “@” and then the binary treatment variable. You should not include a constant in the specification.
Options
coef=arg | Specify the name of the coefficient vector. The default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
did asmrs @ post
estimates an equation by difference-in-difference with ASMRS as the outcome variable, and POST as the treatment variable.
did lemp lpop @ treated
estimates an equation by difference-in-difference with LEMP as the outcome variable, TREATED as the treatment variable, and LPOP as an exogenous regressor.
Cross-references
See
“Difference-in-Difference Estimation” for a discussion of difference-in-difference models.
Execute without opening window.
Syntax
do procedure
do is most useful in EViews programs where you wish to run a series of commands without opening windows in the workfile area.
Examples
output(t) c:\result\junk1
do gdp.adf(c, 4, p)
The first line redirects table output to a file on disk. The second line carries out a unit root test of GDP without opening a window, and prints the results to the disk file.
Cross-references
See
“Object Commands” for a discussion of working with objects.
“Object View and Procedure Reference” provides a complete listing of the views of the various objects.
Convert the entire DRI Basic Economics database into an EViews database.
You must create an EViews database to store the converted DRI data before you use this command. This command may be very time-consuming.
Syntax
driconvert db_name
Follow the command by listing the name of an existing EViews database into which you would like to copy the DRI data. You may include a path name to specify a database not in the default path.
Examples
dbcreate dribasic
driconvert dribasic
driconvert c:\mydata\dridbase
The first line creates a new (empty) database named DRIBASIC in the default directory. The second line copies all the data in the DRI Basic Economics database into in the DRIBASIC database. The last example copies the DRI data into the database DRIDBASE that is located in the C:\MYDATA directory.
Cross-references
See
“EViews Databases” for a discussion of EViews databases.
See also
dbcreate and
db.
Estimation of an elastic net model, including options for Lasso and ridge regression.
Syntax
enet(options) y x1 [x2 x3 ...] [@vw(...)]
List the dependent variable first, followed by a list of the independent variables. Use a “C” if you wish to include an unpenalized intercept term.
Note that PDL and ARMA terms are not permitted in elastic net specifications.
If you wish to specify regressors with an individual penalty weight
, or to place inequality restrictions on the coefficient values, you may do so using special expressions of the form:
@vw(series_name, weight_value)
or
@vw(series_name[, wgt=weight_value, cmin=coef_min, cmax=coef_max])
where weight_value is a non-negative value, coef_min is a non-positive minimum coefficient value, and coef_max is a non-negative maximum coefficient value.
There are two forms of the special expression.
In the abbreviated form, you specify the variable name followed by the penalty weight value.
In the more general form, you specify the variable name followed by one or more keyword expressions in arbitrary order, with the “wgt=” argument specifying the penalty weight, “cmin=” with a non-positive minimum coefficient value, and “cmax=” with a non-negative maximum coefficient value.
When specifying individual regressor behavior using @vw, keep in mind that:
• The special intercept variable “C” is always non-penalized and has an implicit weight
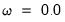
.
• Individual penalty weights may be also specified using a vector in the edit field on the dialog page (or using the command estimation option “lambdawgt=vector_name”). If the vector weights are specified and individual weights are specified using the @vw keyword, the vector weights will be applied first, followed by the individual variable weights.
• Individual coefficient limit values may also be specified using vectors in the and edit fields on the dialog page (or the command estimation options “coefmin=vector_name” and “coefmax=vector_name”). If vector coefficient limits are specified and individual regressor limits are specified using the @vw keyword, the vector limits will be applied first, followed by the individual limits weights.
EViews will normalize the individual penalty weights so that they sum to the number of coefficients.
Options
Specification Options
penalty=arg (default=“el”) | Type of threshold estimation: “enet” (elastic net), “ridge” (ridge), “lasso” (Lasso). |
alpha=arg (default=“.5”) | Value of the mixing parameter. Must be a value from zero to one. |
lambda=arg | Value(s) of the penalty parameter. Can be one or more numbers or vector objects.Values must be zero or greater. If left blank (default) EViews will generate a list. |
Penalty Options
ytrans=arg (default=“none”) | Scaling of the dependent variable: “none” (none), “L1” (L1), “L2” (L2), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
xtrans=arg (default=“stdpop”) | Scaling of the regressor variables: “none” (none), “L1” (L1 norm), “L2” (L2 norm), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
nlambdas=integer (default=100) | Number of penalty values for EViews-supplied list. |
lambdaratio=arg | Ratio of minimum to maximum lambda for EViews-supplied list. You may specify a value for the ratio parameter, or you may leave the edit field blank to let EViews specify a default value based on the number of observations 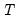 and the number of potential regressors 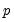 . By default, EViews will set the ratio to 0.001. |
lambdawgt= vector_name | Vector of individual penalty weights, containing non-negative values sized to and matching the order of the variables in the specification. If a vector is provided and individual weights are specified using one or more @vw regressors, the vector weights will be applied first, then overwritten by the individual variable weights. For comparability purposes, we normalize the final weights so that they sum to 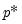 where 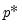 the number of non-zero  . |
nlambdamin=integer (default=5) | Minimum number of lambda values in the path before applying stopping rules. |
minddev=arg (default=1e-05) | Minimum change in deviance fraction to continue estimation. Truncate path estimation if relative change in deviance is smaller than this value. |
maxedev=arg (default=0.99) | Maximum of deviance explained fraction attained to terminate estimation. Truncate path estimation if fraction of null deviance explained is larger than this value. |
maxvars=arg | Maximum number of regressors in the model. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) reaches this value. |
maxvarsratio=arg | Maximum number of regressors in the model as a fraction of the number of observations. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) divided by the number of observations reaches this value. |
Cross Validation Options
cvmethod=arg (default=“kfold_cv”) | Cross-validation method: “kfold” (k-fold), “simple” (simple split), “mcarlo” (Monte Carlo), “leavepout” (leave-P-out), “leave1out” (leave-1-out), “rolling” (rolling window), “expanding” (expanding window). |
cvmeasure=arg (default=“mse”) | Cross-validation fit measure: “mse” (mean-squared error), “r2” (R‑squared), “mae” (mean absolute error), “mape” (mean absolute percentage error), “smape” (symmetric mean absolute percentage error). |
cvnfolds=arg (default=5) | Number of folds for K-fold cross-validation. For “cvmethod=kfold”. |
cvftrain=arg (default=0.8) | Proportion of data for split and Monte Carlo methods. For “cvmethod=simple” and “cvmethod=mcarlo”. |
cvnreps=arg (default=1) | Number of Monte Carlo method repetitions. For “cvmethod=mcarlo”. |
cvleaveout=arg (default=2) | Number of data points left out for leave-p-out method. For “cvmethod=leavepout”. |
cvnwindows=arg (default=4) | Number of windows for rolling window cross-validation method. For “cvmethod=rolling”. |
cvinitial=arg (default=12) | Number of initial data points in the training set for expanding cross-validation. For “cvmethod=expanding”. |
cvpregap=arg (default=0) | Number of observations between end of training set and beginning of test set. For “cvmethod=simple”, “cvmethod=rolling” and “cvmethod=expanding”. |
cvhorizon=arg (default=1) | Number of observation in the test set. For “cvmethod=rolling” and “cvmethod=expanding”. |
cvpostgap=arg (default=0) | Number of observations between end of test set and beginning of next training set for rolling window or between end of test set and end of next training set for expanding window. For “cvmethod=rolling” and “cvmethod=expanding” |
Random Number Options
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
rnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
Other Options
coefmin= vector_name, number | Vector of individual coefficient minimum values, containing negative or missing values sized to and matching the order of the variables in the specification, or a negative value for the minimum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual minimums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
coefmax= vector_name, number | Vector of individual coefficient maximum values, containing positive or missing values sized to and matching the order of the variables in the specification, or a positive value for the maximum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual maximums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
estmethod=arg | Estimation method: “cov” (use covariance algorithm) or “naive” (use naive algorithm). Default is EViews automatic. |
showopts / ‑showopts | [Do / do not] display estimation options in the output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic results view after estimation. |
Examples
The command
enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
estimates an elastic net model with
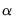
equal to the default of 0.9, no regressor or dependent variable scaling, automatically determined 100-element lambda path with minimum lambda of 0.0001 times the maximum value, using the default K-fold cross-validation with 5 folds with an MSE objective and a random generator seed of 513255899 to determine the optimal value.
Similarly,
enet(penalty=lasso, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
estimates a Lasso model with regressor population standard deviation scaling, with the remaining settings as before, while
enet(penalty=ridge, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
estimates the equivalent ridge regression specification.
The command
enet(alpha=0.75, lambdaratio=.0001, cvmethod=rolling, cvmeasure=smape) lpsa c lcavol lweight age_s lbph svi lcp gleason pgg45
estimates an elastic net model with
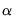
equal to the 0.75 using rolling window cross-validation and SMAPE cross-validation.
We may use the @vw specifications to assign individual penalties and coefficient restrictions
enet(alpha=0.75, lambdaratio=.0001, cvmethod=rolling, cvseed=513255899) lpsa c @vw(lcavol, cmax=.4) lweight age lbph svi lcp gleason @vw(pgg45, cmax=0.075, w=1.2)
estimates an elastic net model with the coefficient of LCAVOL restricted to be less than or equal to 0.4, and the coefficient of PGG45 having a relative penalty weight of 1.2, and a maximum value of 0.075.
Identical specifications may be estimated using vectors of penalty weights and coefficient restrictions,
vector(9) cmax = na
cmax(2) = 0.4
cmax(9) = 1.2
vector(9) lwgt = 1
lwgt(9) = 1.2
enet(alpha=0.75, coefmax=cmax, lambdawgt=lwgt, lambdaratio=.0001, cvmethod=rolling, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
and
vector(9) cmax = na
cmax(2) = 0.4
vector(9) lwgt = 1
lwgt(9) = 50
enet(alpha=0.75, coefmax=cmax, lambdawgt=lwgt, lambdaratio=.0001, cvmethod=rolling, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason @vw(pgg45, cmax=0.075, w=1.2)
since the penalty weight for PGG45 in the vector is overwritten by the individual weight specified using the @vw.
Note that in neither case is the intercept penalized, even though the corresponding element of LWGT is equal to 1 since the specification of “C” is always implicitly treated as “@VW(C, 0)”.
Cross-references
See
“Elastic Net and Lasso” for a discussion of elastic net, ridge regression, and Lasso models.
See
Equation::enet for the object version of this command.
Unregister a program file as an EViews Add-in.
Syntax
deleteaddin(options) [path\]prog_name
unregisters the specified program file as an EViews Add-in.
If you do not provide the optional path specification, EViews looks for the program file in the default EViews Add-ins directory.
Explicit path specifications containing “.\” and “..\” (to indicate the current level and one directory level up) are evaluated relative EViews default directory.
You may use the special “<addins>” directory keyword in your path specification.
Options
type=arg | Specify the Add-ins type, where arg is the name of a EViews object type. The default is to create a global Add-in. |
proc=arg | User-defined command/procedure name. If omitted, the Add-in will not have a command form. |
Examples
deleteaddin .\myaddin.prg
unregisters the Add-in associated with file “Myaddin.prg”.
Alternatively,
deleteaddin(proc="myaddin")
unregisters the Add-in whose proc name matches “myaddin”. Note that this name may not match the program name.
deleteaddin(type="graph", proc="recshade")
unregisters the graph “Recshade” specific Add-in. In cases, where more than 1 Add-in has the same proc name, the type is useful to differentiate which is to be unregistered.
Cross-references
See
“Add-ins” for a detailed discussion of Add-ins.
Execute a program.
The exec command executes a program. The program may be located in memory or stored in a program file on disk.
Syntax
exec(options) [path\]prog_name(prog_options) [%0 %1 …]
If you wish to pass one or more options to the program, you should enclose them in parentheses immediately after the filename. If the program has arguments, you should list them after the filename.
EViews first checks to see if the specified program is in memory. If the program is not located, EViews then looks for the program on disk in the EViews Add-ins directory, or in the specified path. The program file should have a “.PRG” extension, which you need not specify in the prog_name.
Options
integer (default=1) | Set maximum errors allowed before halting the program in interactive mode. Note that the integer option does not apply when using exec in a program, it only applies when using exec from the command line. When using exec in a parent program to execute a child program, the child program inherits the maximum error count from the parent. |
c | Run program file without opening a window for display of the program file. |
verbose / quiet | Verbose mode in which messages will be sent to the status line at the bottom of the EViews window (slower execution), or quiet mode which suppresses workfile display updates (faster execution). |
v / q | Same as [verbose / quiet]. |
ver4 / ver5 | Execute program in [version 4 / version 5] compatibility mode. |
this=object_name | Set the _this object for the executed program. If omitted, the executed program will inherit the _this object from the parent program, or from the current active workfile object when the exec command is issued from the command window. |
Examples
exec rollreg
will run the program “Rollreg.prg” in the EViews add-in directory.
exec(this=graph01) recshade
will run the program “Recshade” in the EViews add-in directory, setting the _this object to GRAPH01.
exec(4) c:\myfiles\simul.prg(h=3) xhat
will run the program “Simul.prg” in the path “c:\myfiles\”, with program option string “h=3”, the %0 argument set to “XHAT”, and with the maximum error count set to 4.
Note that in contrast to the run command, exec will not stop executing a running program after returning from the executed program. For example if you have a program containing:
exec simul
print x
the print statement will be executed after running the “Simul.prg” program. If you replace exec with run, the program will stop after executing the commands in “Simul.prg”.
Cross-references
See also
run and
include.
Exit from EViews (close the EViews application).
You will be prompted to save objects and workfiles which have changed since the last time they were saved to disk. Be sure to save your workfiles, if desired, since all changes that you do not save to a disk file will be lost.
Syntax
exit
Cross-references
Expand a workfile.
No longer supported. See the replacement command
pagestruct.
Factor breakpoint test for stability.
Carries out a factor breakpoint test for parameter constancy.
Syntax
facbreak(options) ser1 [ser2 ser3 ...] @ x1 x2 x3
You must provide one or more series to be used as the factors with which to split the sample into categories. To specify more than one factor, separate the factors by a space. If the equation is specified by list and contains no linear terms, you may specify a subset of the regressors to be tested for a breakpoint after an “@” sign.
Options
p | Print the result of the test. |
Examples
The commands
ls log(spot) c log(p_us) log(p_uk)
facbreak season
perform a regression of the log of SPOT on a constant, the log of P_US, and the log of P_UK, and employ a factor breakpoint test to determine whether the parameters are stable through the different values of SEASON.
To test whether only the constant term and the coefficient on the log of P_US are “stable” enter the commands:
facbreak season @ c log(p_us)
Cross-references
See
“Factor Breakpoint Test” for further discussion.
Specify and estimate a factor analysis model.
Syntax
factest(method=arg, options) x1 [x2 x3...] [@partial z1 z2 z3...]
factest(method=arg, options) matrix_name [[obs] [conditioning]] [@ name1 name2 name3...]
where:
method=arg (default= “ml”) | Factor estimation method: “ml” (maximum likelihood), “gls” (generalized least squares), “ipf” (iterated principal factors), “pace” (non-iterative partitioned covariance matrix estimation), “pf” (principal factors), “uls” (unweighted least squares) |
and the available options are specific to the factor estimation method (see
“Factor Methods”).
The factest command allows you to estimated a factor analysis model without first declaring a factor object and then applying an estimation method. It provides a convenient method of interactively estimating transitory specifications that are not to be named and saved with the workfile.
Estimation of a factor analysis specification using
factest only differs from estimation using a named factor and a factor estimation procedure (e.g.,
Factor::ipf) in the use of the “method=” option and in the fact that the command results in an unnamed factor object.
Examples
The command:
factest(method=gls) g1
estimates a factor analysis model for the series in G1 using GLS. The result is an unnamed factor object. (Almost) equivalently, we may declaring and estimate the factor analysis object using the
Factor::gls estimation method procedure
factor f1.gls g1
which differs only in the fact that the former yields an unnamed factor object and the latter saves the object F1 in the workfile.
The command:
factest(method=ml) group01 @partial ser1 ser2
estimates the factor model using the partial correlation for the series in GROUP01, conditional on the series SER1 and SER2. The command is equivalent to:
factor f2.ml group01 @partial ser1 ser2
except the latter names the factor object F2.
Cross-references
See
“Factor Analysis” for a general discussion of factor analysis. The various estimation methods are described in
“Estimation Methods”.
Fetch objects from databases or databank files into the workfile.
fetch reads one or more objects from EViews databases or databank files into the active workfile. The objects are loaded into the workfile using the object in the database or using the databank file name.
If you fetch a series into a workfile with a different frequency, EViews will automatically apply the frequency conversion method attached to the series by setconvert. If the series does not have an attached conversion method, EViews will use the method set by in the main menu. You can override the conversion method by specifying an explicit conversion method option.
Syntax
fetch(options) object_list
The fetch command keyword is followed by a list of object names separated by spaces. The default behavior is to fetch the objects from the default database (this is a change from versions of EViews prior to EViews 3.x where the default was to fetch from individual databank files).
You can precede the object name with a database name and the double colon “::” to indicate a specific database source. If you specify the database name as an option in parentheses (see below), all objects without an explicit database prefix will be fetched from the specified database. You may optionally fetch from individual databank files or search among registered databases.
You may use wild card characters, “?” (to match a single character) or “*” (to match zero or more characters), in the object name list. All objects with names matching the pattern will be fetched.
To fetch from individual databank files that are not in the default path, you should include an explicit path. If you have more than one object with the same file name (for example, an equation and a series named CONS), then you should supply the full object file name including identifying extensions.
Options
d=db_name | Fetch from specified database. |
d | Fetch all registered databases in registry order. |
i | Fetch from individual databank files. |
link | Fetch as a database link. |
notifyillegal | When in a program, report illegal EViews object names. By default, objects with illegal names are automatically renamed. (Has no effect in the command window.) |
p=prefix | Specify a naming prefix that will be prepended to the listed names to be fetched. |
s=suffix | Specify a naming suffix that will be appended to the listed names to be fetched. |
pi=prefix | Specify a naming prefix that will be removed from the name of the source objects once fetched into the workfile. |
si=suffix | Specify a naming suffix that will be removed from the name of the source objects once fetched into the workfile. |
tr=integer | Truncate long series names to integer characters. The default value of integer is 24. |
The following options are available for fetch of group objects:
g=arg | Group fetch options: “b” (fetch both group definition and series), “d” (fetch only the series in the group), “l” (fetch only the group definition). |
The database specified by the double colon “::” takes precedence over the database specified by the “d=” option.
The following options control the frequency conversion method when copying series and group objects into a workfile page and converting from low to high frequency:
c=arg | Low to high conversion methods: “r” or “repeata” (constant match average), “d” or “repeats” (constant match sum), “q” or “quada” (quadratic match average), “t” or “quads” (quadratic match sum), “linearf” (linear match first), “i” or “linearl” (linear match last), “cubicf” (cubic match first), “c” or “cubicl” (cubic match last), “pointf” (point first), “pointl” (point last). |
The following options control the frequency conversion method when copying series and group objects to a workfile, converting from high to low frequency:
c=arg | High to low conversion methods removing NAs: “a” (average of the nonmissing observations), “s” (sum of the nonmissing observations), “f” (first nonmissing observation), “l” (last nonmissing observation), “x” (maximum nonmissing observation), “m” (minimum nonmissing observation). High to low conversion methods propagating NAs: “an” or “na” (average, propagating missings), “sn” or “ns” (sum, propagating missings), “fn” or “nf” (first, propagating missings), “ln” or “nl” (last, propagating missings), “xn” or “nx” (maximum, propagating missings), “mn” or “nm” (minimum, propagating missings). |
If no conversion method is specified, the series-specific or global default conversion method will be employed.
Examples
To fetch M1, GDP, and UNEMP from the default database, use:
fetch m1 gdp unemp
To fetch M1 and GDP from the US1 database and UNEMP from the MACRO database, use the command:
fetch(d=us1) m1 gdp macro::unemp
You can fetch all objects with names starting with “SP” by searching all registered databases in the search order. The “c=f” option uses the first (nonmissing) observation to convert the frequency of any matching series with a higher frequency than the destination workfile frequency:
fetch(d,c=f) sp*
You can fetch M1 and UNEMP from individual databank files using:
fetch(i) m1 c:\data\unemp
To fetch all objects with names starting with “CONS” from the two databases USDAT and UKDAT, use the command:
fetch usdat::cons* ukdat::cons*
The command:
fetch a?income
will fetch all series beginning with the letter “A”, followed by any single character, and ending with the string “income”.
Use the “notifyillegal” option to display a dialog when fetching the series MYILLEG@LNAME that will suggest a valid name and give you to opportunity to name the object before it is inserted into a workfile:
fetch(notifyillegal) myilleg@lname
The command:
fetch(p=us_) gdp inv cons
Fetches the series US_GDP, US_INV and US_CONS into workfile series with the same names.
fetch(si=” us equity”) ibm msft
Fetches the database series IBM US EQUITY, and MSFT US EQUITY into workfile series with names IBM and MSFT.
Specifying the “tr” option causes EViews 10 to truncate long series names to 24 characters (by default) instead of 300. This will let programs written for EViews 9 continue to work with EViews 10:
fetch(d=eia, link, tr) coal.average_employees.al-tot.a
The “tr” option can also be set to a number between 0 and 300:
fetch(d=eia, link, tr=20) coal.average_employees.al-tot.a
Cross-references
See
“EViews Databases” for a discussion of databases, databank files, and frequency conversion.
Appendix A. “Wildcards” describes the use of wildcard characters.
See also
store, and
copy.
See
Series::setconvert for information on default conversion settings.
Computes static forecasts or fitted values from an estimated equation.
When the regressor contains lagged dependent values or ARMA terms, fit uses the actual values of the dependent variable instead of the lagged fitted values. You may instruct fit to compare the forecasted data to actual data, and to compute forecast summary statistics.
(Note that we recommend that you instead use the equation proc
Equation::fit since it explicitly specifies the equation of interest.)
Not available for equations estimated using ordered methods; use
Equation::makemodel to create a model using the ordered equation results.
Syntax
fit(options) yhat [y_se]
Following the fit keyword, you should type a name for the forecast series and, optionally, a name for the series containing the standard errors and, for ARCH specifications, a name for the conditional variance series.
Forecast standard errors are currently not available for binary, censored, and count models.
Basic Options
d | In models with implicit dependent variables, forecast the entire expression rather than the normalized variable. |
u | Substitute expressions for all auto-updating series in the equation. |
g | Graph the fitted values together with the ±2 standard error bands. |
ga | Graph the forecasts along with the actuals (if available). |
e | Produce the forecast evaluation table. |
i | Compute the fitted values of the index. Only for binary, censored and count models. |
s | Ignore ARMA terms and use only the structural part of the equation to compute the fitted values. |
n | Ignore coef uncertainty in standard error calculations that use them. |
forcsmpl = smpl | Fit sample (optional). If forecast sample is not provided, the workfile sample will be employed. |
f = arg (default= “actual”) | Out-of-fit-sample fill behavior: “actual” (fill observations outside the fit sample with actual values for the fitted variable), “na” (fill observations outside the fit sample with missing values). |
prompt | Force the dialog to appear from within a program. |
p | Print view. |
Stochastic Options
Options for forecasting from a functional coefficients estimated equation.
stochastic = arg (default = “none”) | Stochastic method: “none” (none), “mca” (Monte Carlo –asymptotic), “mcbs” (Monte Carlo – bootstrap), “bs” (bootstrap). |
reps = integer (default = 999) | Number of stochastic replications |
lhr = arg (default = 0.1) | Lower historical range (number between 0 and upper historical range). |
uhr = arg (default = 0.9) | Upper historical range (number between lower historical range and 1). |
bsdep | Bootstrap only the dependent variable (not the functional coefficient variable). |
Examples
equation eq1.ls cons c cons(-1) inc inc(-1)
fit c_hat c_se
genr c_up=c_hat+2*c_se
genr c_low=c_hat-2*c_se
line cons c_up c_low
The first line estimates a linear regression of CONS on a constant, CONS lagged once, INC, and INC lagged once. The second line stores the static forecasts and their standard errors as C_HAT and C_SE. The third and fourth lines compute the +/- 2 standard error bounds. The fifth line plots the actual series together with the error bounds.
equation eq2.binary(d=l) y c wage edu
fit yf
fit(i) xbeta
genr yhat = 1-@clogit(-xbeta)
The first line estimates a logit specification for Y with a conditional mean that depends on a constant, WAGE, and EDU. The second line computes the fitted probabilities, and the third line computes the fitted values of the index. The fourth line computes the probabilities from the fitted index using the cumulative distribution function of the logistic distribution. Note that YF and YHAT should be identical.
Note that you cannot fit values from an ordered model. You must instead solve the values from a model. The following lines generate fitted probabilities from an ordered model:
equation eq3.ordered y c x z
eq3.makemodel(oprob1)
solve oprob1
The first line estimates an ordered probit of Y on a constant, X, and Z. The second line makes a model from the estimated equation with a name OPROB1. The third line solves the model and computes the fitted probabilities that each observation falls in each category.
Cross-references
To perform dynamic forecasting, use
forecast.
See
“Forecasting from an Equation” for a discussion of forecasting in EViews and
“Discrete and Limited Dependent Variable Models” for forecasting from binary, censored, truncated, and count models.
See
“Forecasting” for a discussion of forecasting from sspace models.
See
Equation::forecast and
Equation::fit for the equivalent object commands.
See
Equation::makemodel and
Model::solvefor forecasting from systems of equations or ordered equations.
Computes dynamic forecasts of the default equation.
forecast computes the forecast using the default equation for all observations in a specified sample. In some settings, you may instruct forecast to compare the forecasted data to actual data, and to compute summary statistics.
(Note that we recommend that you instead use the equation proc
Equation::forecast since it explicitly specifies the equation of interest.)
Syntax
forecast(options) yhat [y_se]
You should enter a name for the forecast series and, optionally, a name for the series containing the standard errors. Forecast standard errors are currently not available for binary or censored models. forecast is not available for models estimated using ordered methods.
Options
d | In models with implicit dependent variables, forecast the entire expression rather than the normalized variable. |
u | Substitute expressions for all auto-updating series in the equation. |
g | Graph the forecasts together with the ±2 standard error bands. |
e | Produce the forecast evaluation table. |
i | Compute the forecasts of the index. Only for binary, censored and count models. |
s | Ignore ARMA terms and use only the structural part of the equation to compute the forecasts. |
n | Ignore coef uncertainty in standard error calculations that use them. |
b =arg | MA backcast method: “fa” (forecast available). Only for equations estimated with MA terms. This option is ignored if you specify the “s” (structural forecast) option. The default method uses the estimation sample. |
f = arg (default= “actual”) | Out-of-forecast-sample fill behavior: “actual” (fill observations outside the forecast sample with actual values for the fitted variable), “na” (fill observations outside the forecast sample with missing values). |
stochastic | Perform stochastic simulation for dynamic equations estimated using least squares. |
streps=integer (default=1000) | Number of stochastic repetitions (for threshold regression or stochastic simulation). |
stfrac=number (default=.02) | Fraction of failed repetitions before stopping (for threshold regression or stochastic simulation). |
prompt | Force the dialog to appear from within a program. |
p | Print view. |
Examples
The following lines:
smpl 1970q1 1990q4
equation eq1.ls con c con(-1) inc
smpl 1991q1 1995q4
forecast con_d
plot con_d
estimate a linear regression over the period 1970Q1–1990Q4, computes dynamic forecasts for the period 1991Q1–1995Q4, and plots the forecast as a line graph.
equation eq1.ls m1 gdp ar(1) ma(1)
forecast m1_bj bj_se
forecast(s) m1_s s_se
plot bj_se s_se
estimates an ARMA(1,1) model, computes the forecasts and standard errors with and without the ARMA terms, and plots the two forecast standard errors.
Cross-references
To perform static forecasting, see
fit.
See
“Forecasting from an Equation” for a discussion of forecasting in EViews and
“Discrete and Limited Dependent Variable Models” for forecasting from binary, censored, truncated, and count models.
See
“Forecasting” for a discussion of forecasting from sspace models.
See
Equation::forecast and
Equation::fit for the equivalent object commands.
See
Equation::makemodel and
Model::solvefor forecasting from systems of equations or ordered equations.
Creates graph, table, or text objects from a view.
Syntax
freeze(options, name) object_name.view_command
If you follow the keyword freeze with an object name but no view of the object, freeze will use the default view for the object. You may provide a destination name for the object containing the frozen view in parentheses.
Options
mode = overwrite | Overwrites the object name if it already exists. |
Examples
freeze gdp.uroot(4,t)
creates an untitled table that contains the results of the unit root test of the series GDP.
group rates tb1 tb3 tb6
freeze(gra1) rates.line(m)
show gra1.align(2,1,1)
freezes a graph named GRA1 that contains multiple line graphs of the series in the group RATES, realigns the individual graphs, and displays the resulting graph.
freeze(mygra) gra1 gra3 gra4
show mygra.align(2,1,1)
creates a graph object named MYGRA that combines three graph objects GRA1, GRA2, and GRA3, and displays MYGRA in two columns.
freeze(mode=overwrite, mygra) gra1 gra2 gra3
show mygra.align(2,1,1)
creates a graph object MYGRA that combines the three graph objects GRA1, GRA2 and GRA3, and displays MYGRA in two columns. If the object MYGRA already exists, it would be replaced by the new object.
Cross-references
See
“Object Commands” for discussion. See also
“Object Basics” for further discussion of objects and views of objects.
Freezing graph views is described in
“Creating Graph Objects”.
Declare a series object with a formula for auto-updating, or specify a formula for an existing series.
Syntax
frml series_name = series_expression
frml series_name = @clear
Follow the frml keyword with a name for the object, and an assignment statement. The special keyword “@CLEAR” is used to return the auto-updating series to an ordinary numeric or alpha series.
Examples
To define an auto-updating numeric or alpha series, you must use the frml keyword prior to entering an assignment statement. The following example creates a series named LOW that uses a formula to compute its values.:
frml low = inc<=5000 or edu<13
The auto-updating series takes the value 1 if either INC is less than or equal to 5000 or EDU is less than 13, and 0 otherwise, and will be re-evaluated whenever INC or EDU change.
If FIRST_NAME and LAST_NAME are alpha series, then the formula declaration:
frml full_name = first_name + " " + last_name
creates an auto-updating alpha series FULL_NAME.
You may apply a frml to an existing series. The commands:
series z = 3
frml z =(x+y)/2
makes the previously created series Z an auto-updating series containing the average of series X and Y. Note that once a series is defined to be auto-updating, it may not be modified directly. Here, you may not edit Z, nor may you generate values into the series.
Note that the commands:
series z = 3
z = (x+y)/2
while similar, produce quite different results, since the absence of the frml keyword in the second example means that EViews will generate fixed values in the series instead of defining a formula to compute the series values. In this latter case, the values in the series Z are fixed, and may be modified.
One particularly useful feature of auto-updating series is the ability to reference series in databases. The command:
frml gdp = usdata::gdp
creates a series called GDP that obtains its values from the series GDP in the database USDATA. Similarly:
frml lgdp = log(usdata::gdp)
creates an auto-updating series that is the log of the values of GDP in the database USDATA.
To turn off auto-updating for a series or alpha, you should use the special expression “@CLEAR” in your frml assignment. The command:
frml z = @clear
sets the series to numeric or alpha value format, freezing the contents of the series at the current values.
Cross-references
See
“Auto-Updating Series” for a discussion of updating series.
Estimate a functional coefficient regression equation.
Syntax
funcoef(options) y x1 [x2 x3 ...] @ funcoef_series
List the funcoef keyword, the dependent variable and a list of the regressor variables, followed by the “@” symbol and the name of the functional coefficient series.
Options
Basic Options
kern=arg (default=“epan”) | Kernel type: “epan” (Epanechnikov, default), “trngl” (Triangular), “unif” (Uniform), “gauss” (Normal–Gaussian), “bi” (Biweight–Quartic), “tri” (Triweight). |
eval=arg (default=“data”) | Evalution points: observed data (“data”), grid of values (“grid”). if “eval=grid” you must specify the grid values using “gmin=”, “gmax=” and “glen=”, or using “gvec=”. |
gmin = arg | Estimation grid minimum (if “eval=grid”). Must be specified along with “gmax=” and “glen=”. |
gmax = arg | Estimation grid maximum (if “eval=grid”). Must be specified along with “gmin=” and “glen=”. |
glen = arg | Estimation grid length (if “eval=grid”). Must be specified along with “gmin=” and “gmax=”. |
gvec = arg | Estimation grid points in a vector object (if “eval=grid”). |
plyk = arg (default = 1) | Estimation polynomial degree for final stage. |
auxk = arg (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage. |
p | Print results. |
Pilot Bandwidth Options
plth =arg (default = “rsc”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
Final Bandwidth Options
fnlh =arg (default = “cv”) | Final bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), integrated asymptotic mean square error (“mse”), leave-one-out cross-validation (“loo”), nonparametric AIC (“aic”), user-defined (“user”). |
fnlbw=arg | User-defined bandwidth (if “fnlh=user”). |
fnlhmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “fnlh=user”). |
fnlhmax=arg (default =1) | Bandwidth grid search maximum value (if not “fnlh=user”). |
fnlhlen=integer (default = 100) | Bandwidth grid search length (if not “fnlh=user”). |
fnlhinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “fnlh=user”). |
fnlhcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (if not “fnlh=user”). |
fnlm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “fnlh=cv”). |
fnlfq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “fnlh=cv”). |
Examples
We consider examples for three equations that estimate FCOEF using UNRATE as the dependent variable, UNRATE(-1 to -2) as independent variables, and LWAGE(-4) as the functional coefficient variable.
funcoef(eval=grid, gmin=0, gmax=10, glen=100) unrate unrate(-1) unrate(-2) @ lwage(-4)
evaluates over a custom uniform grid from 0 to 10 with length 100.
funcoef(eval=grid, gvec=vecgrid) unrate unrate(-1) unrate(-2) @ lwage(-4)
evaluates over a custom grid provided by the values of a workfile vector called VECGRID.
funcoef(plyk=3, auxk=5) unrate unrate(-1) unrate(-2) @ lwage(-4)
estimates using local polynomial fitting with main polynomial degree 3 and auxiliary polynomial degree 5. The latter is employed deriving bias, variance, and bandwidths.
Cross-references
See
“Functional Coefficient Regression” for additional discussion on functional coefficients estimation.
Generate series.
Generate series or alphas.
Syntax
genr ser_name = expression
Examples
genr y = 3 + x
generates a numeric series that takes the values from the series X and adds 3.
genr full_name = first_name + last_name
creates an alpha series formed by concatenating the alpha series FIRST_NAME and LAST_NAME.
Cross-references
See
“Working with Data” for discussion of generating series data.
See
Series::series and
Alpha::alpha for a discussion of the expressions allowed in
genr.
Estimate a Generalized Linear Model (GLM).
Syntax
glm(options) spec
List the glm keyword, followed by the dependent variable and a list of the explanatory variables, or an explicit linear expression.
If you enter an explicit linear specification such as “Y=C(1)+C(2)*X”, the response variable will be taken to be the variable on the left-hand side of the equality (“Y”) and the linear predictor will be taken from the right-hand side of the expression (“C(1)+C(2)*X”).
Offsets may be entered directly in an explicit linear expression or they may be entered as using the “offset=” option.
Specification Options
family=arg (default=“normal”) | Distribution family: Normal (“normal”), Poisson (“poisson”), Binomial Count (“binomial”), Binomial Proportion (“binprop”), Negative Binomial (“negbin”), Gamma (“gamma”), Inverse Gaussian (“igauss”), Exponential Mean (“emean)”, Power Mean (“pmean”), Binomial Squared (“binsq”). The Binomial Count, Binomial Proportion, Negative Binomial, and Power Mean families all require specification of a distribution parameter: |
n=arg (default=1) | Number of trials for Binomial Count (“family=binomial”) or Binomial Proportions (“family=binprop”) families. |
fparam=arg | Family parameter value for Negative Binomial (“family=negbin”) and Power Mean (“family=pmean”) families. |
link=arg (default=“identity”) | Link function: Identity (“identity”), Log (“log”), Log Compliment (“logc”), Logit (“logit”), Probit (“probit”), Log-log (“loglog”), Complementary Log-log (“cloglog”), Reciprocal (“recip”), Power (“power”), Box-Cox (“boxcox”), Power Odds Ratio (“opow”), Box-Cox Odds Ratio (“obox”). The Power, Box-Cox, Power Odds Ratio, and Box-Cox Odds Ratio links all require specification of a link parameter specified using “lparam=”. |
lparam=arg | Link parameter for Power (“link=power”), Box-Cox (“link=boxcox”), Power Odds Ratio (“link=opow”) and Box-Cox Odds Ratio (“link=obox”) link functions. |
offset=arg | Offset terms. |
disp=arg | Dispersion estimator: Pearson 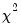 statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”). The default is family specific: “unit” for Binomial Count, Binomial Proportion, Negative Binomial, and Poison, and “pearson” for all others. The “deviance” option is only offered for families in the exponential family of distributions (Normal, Poisson, Binomial Count, Binomial Proportion, Negative Binomial, Gamma, Inverse Gaussian). |
dispval=arg | User-dispersion value (if “disp=user”). |
fwgts=arg | Frequency weights. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
In addition to the specification options, there are options for estimation and covariance calculation.
Additional Options
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “fisher” (IRLS – Fisher Scoring), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
estmeth=arg (default=”marquardt”) | Legacy estimation algorithm: Quadratic Hill Climbing (“marquardt”), Newton-Raphson (“newton”), IRLS - Fisher Scoring (“irls”), BHHH (“bhhh”). (Applicable when “optmethod=legacy”.) |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
preiter=arg (default=0) | Number of IRLS pre-iterations to refine starting values (only available for non-IRLS estimation). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “glm” (GLM method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “fisher” (expected Hessian). (Applicable when “optmethod=” not equal to “legacy”. |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). Applicable where “cov=hac”. |
covinfosel=arg (default=”aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). For settings where “cov=hac, covlag=a”. |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 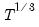 . For settings where “cov=hac, covlag=a”. |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). For settings where “cov=hac”. |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). For settings where “cov=hac” and “covkern=” is specified. |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). For settings where “cov=hac” and “covkern=” is specified. |
covbwoffset=number | Apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
covbwint | Use integer portion of kernel bandwidth. For settings where “cov=hac” and “covkern=” is specified. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
glm(link=log) numb c ip feb
estimates a normal regression model with exponential mean.
glm(family=binomial, n=total) disease c snore
estimates a binomial count model with default logit link where TOTAL contains the number of binomial trials and DISEASE is the number of binomial successes. The specification
glm(family=binprop, n=total, cov=huber, nodf) disease/total c snore
estimates the same specification in proportion form, and computes the coefficient covariance using the Huber-White sandwich with no d.f. correction.
glm(family=binprop, disp=pearson) prate mprate log(totemp) log(totemp)^2 age age^2 sole
estimates a binomial proportions model with default logit link, but computes the coefficient covariance using the GLM scaled covariance with dispersion computed using the Pearson Chi-square statistic.
glm(family=binprop, link=probit, cov=huber) prate mprate log(totemp) log(totemp)^2 age age^2 sole
estimates the same basic specification, but with a probit link and Huber-White standard errors.
glm(family=poisson, offset=log(pyears)) los hmo white type2 type3 c
estimates a Poisson specification with an offset term LOG(PYEARS).
Cross-references
See
Equation::glm for the equivalent equation object command.
Estimation by generalized method of moments (GMM).
Syntax
gmm(options) y x1 [x2 x3...] @ z1 [z2 z3...]
gmm(options) specification @ z1 [z2 z3...]
Follow the name of the dependent variable by a list of regressors, followed by the “@” symbol, and a list of instrumental variables which are orthogonal to the residuals. Alternatively, you can specify an expression using coefficients, an “@” symbol, and a list of instrumental variables. There must be at least as many instrumental variables as there are coefficients to be estimated.
In panel settings, you may specify dynamic instruments corresponding to predetermined variables. To specify a dynamic instrument, you should tag the instrument using “@DYN”, as in “@DYN(X)”. By default, EViews will use a set of period-specific instruments corresponding to lags from -2 to “-infinity”. You may also specify a restricted lag range using arguments in the “@DYN” tag. For example, to use lags from-5 to “-infinity” you may enter “@DYN(X, -5)”; to specify lags from -2 to -6, use “@DYN(X, -2, -6)” or “@DYN(X, -6, -2)”.
Note that dynamic instrument specifications may easily generate excessively large numbers of instruments.
Options
Non-Panel GMM Options
Basic GMM Options
nocinst | Do not include automatically a constant as an instrument. |
method=keyword | Set the weight updating method. keyword should be one of the following: “nstep” (N-Step Iterative, or Sequential N-Step Iterative, default), “converge” (Iterate to Convergence or Sequential Iterate to Convergence), “simul” (Simultaneous Iterate to Convergence), “oneplusone” (One-Step Weights Plus One Iteration), or “cue” (Continuously Updating”. |
gmmiter=integer | Number of weight iterations. Only applicable if the “method=nstep” option is set. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR or MA terms (see also
param). |
s=number | Determine starting values for equations specified by list with AR or MA terms. Specify a number between zero and one representing the fraction of TSLS estimates computed without AR or MA terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR and MA terms which are instead set to EViews determined default values. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get the one-step weighting matrix. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Estimation Weighting Matrix Options
instwgt=keyword | Set the estimation weighting matrix type. Keyword should be one of the following: “tsls” (two-stage least squares), “white” (White diagonal matrix), “hac” (Newey-West HAC, default) or “user” (user defined). |
instwgtmat=name | Set the name of the user-defined estimation weighting matrix. Only applicable if the “instwgt=user” option is set. |
instlag=arg (default=1) | Whitening Lag specification: integer (user-specified lag value), “a” (automatic selection). |
instinfosel=arg (default=“aic”) | Information criterion for automatic whitening lag selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “instlag=a”). |
instmaxlag= integer | Maximum lag-length for automatic selection (optional) (if “instlag=a”). The default is an observation-based maximum of 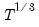 . |
instkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
instbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
instnwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “instbw=neweywest”). |
instbwint | Use integer portion of bandwidth. |
Covariance Options
cov=keyword | Covariance weighting matrix type (optional): “updated” (estimation updated), “tsls” (two-stage least squares), “white” (White diagonal matrix), “hac” (Newey-West HAC), “wind” (Windmeijer) or “user” (user defined). The default is to use the estimation weighting matrix. |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covwgtmat=name | Set the name of the user-definied covariance weighting matrix. Only applicable if the “covwgt=user” option is set. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=”aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 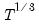 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
Panel GMM Options
cx=arg | Cross-section effects method: (default) none, fixed effects estimation (“cx=f”), first-difference estimation (“cx=fd”), orthogonal deviation estimation (“cx=od”) |
per=arg | Period effects method: (default) none, fixed effects estimation (“per=f”). |
levelper | Period dummies always specified in levels (even if one of the transformation methods is used, “cx=fd” or “cx=od”). |
wgt=arg | GLS weighting: (default) none, cross-section system weights (“wgt=cxsur”), period system weights (“wgt=persur”), cross-section diagonal weighs (“wgt=cxdiag”), period diagonal weights (“wgt=perdiag”). |
gmm=arg | GMM weighting: 2SLS (“gmm=2sls”), White period system covariances (Arellano-Bond 2-step/n-step) (“gmm=perwhite”), White cross-section system (“gmm=cxwhite”), White diagonal (“gmm=stackedwhite”), Period system (“gmm=persur”), Cross-section system (“gmm=cxsur”), Period heteroskedastic (“cov=perdiag”), Cross-section heteroskedastic (“gmm=cxdiag”). By default, uses the identity matrix unless estimated with first difference transformation (“cx=fd”), in which case, uses (Arellano-Bond 1-step) difference weighting matrix. In this latter case, you should specify 2SLS weights (“gmm=2sls”) for Anderson-Hsiao estimation. |
cov=arg | Coefficient covariance method: (default) ordinary, White cross-section system robust (“cov=cxwhite”), White period system robust (“cov=perwhite”), White heteroskedasticity robust (“cov=stackedwhite”), Cross-section system robust/PCSE (“cov=cxsur”), Period system robust/PCSE (“cov=persur”), Cross-section heteroskedasticity robust/PCSE (“cov=cxdiag”), Period heteroskedasticity robust (“cov=perdiag”). |
keepwgts | Keep full set of GLS/GMM weights used in estimation with object, if applicable (by default, only weights which take up little memory are saved). |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
iter=arg (default=“onec”) | Iteration control for GLS and GMM weighting specifications: perform one weight iteration, then iterate coefficients to convergence (“iter=onec”), iterate weights and coefficients simultaneously to convergence (“iter=sim”), iterate weights and coefficients sequentially to convergence (“iter=seq”), perform one weight iteration, then one coefficient step (“iter=oneb”). |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR or MA terms (see also
param). |
s=number | Determine starting values for equations specified by list with AR terms. Specify a number between zero and one representing the fraction of TSLS estimates computed without AR terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR terms which are instead set to EViews determined default values. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get the one-step weighting matrix. |
unbalsur | Compute SUR factorization in unbalanced data using the subset of available observations for a cluster. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
p | Print results. |
Note that some options are only available for a subset of specifications.
Examples
In a non-panel workfile, we may estimate equations using the standard GMM options. The specification:
gmm(instwgt=white,gmmiter=2,nodf) cons c y y(-1) w @ c p(-1) k(-1) x(-1) tm wg g t
estimates the Klein equation for consumption using GMM with a White diagonal weighting matrix (two steps and no degree of freedom correction). The command:
gmm(method=cue,instwgt=hac,instlag=1,instkern=thann, instbw=andrews,nodf) i c y y(-1) k(-1) @ c p(-1) k(-1) x(-1) tm wg g t
estimates the Klein equation for investment using a Newey-West HAC weighting matrix, with pre-whitening with 1 lag, a Tukey-Hanning kernel and the Andrews automatic bandwidth routine. The estimation is performed using continuously updating weight iterations.
When working with a workfile that has a panel structure, you may use the panel equation estimation options. The command
gmm(cx=fd, per=f) dj dj(-1) @ @dyn(dj)
estimates an Arellano-Bond “1-step” estimator with differencing of the dependent variable DJ, period fixed effects, and dynamic instruments constructed using DJ with observation specific lags from period
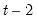
to 1.
To perform the “2-step” version of this estimator, you may use:
gmm(cx=fd, per=f, gmm=perwhite, iter=oneb) dj dj(-1) @ @dyn(dj)
where the combination of the options “gmm=perwhite” and (the default) “iter=oneb” instructs EViews to estimate the model with the difference weights, to use the estimates to form period covariance GMM weights, and then re-estimate the model.
You may iterate the GMM weights to convergence using:
gmm(cx=fd, per=f, gmm=perwhite, iter=seq) dj dj(-1) @ @dyn(dj)
Alternately:
gmm(cx=od, gmm=perwhite, iter=oneb) dj dj(-1) x y @ @dyn(dj,-2,-6) x(-1) y(-1)
estimates an Arellano-Bond “2-step” equation using orthogonal deviations of the dependent variable, dynamic instruments constructed from DJ from period
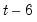
to
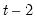
, and ordinary instruments X(-1) and Y(-1).
Cross-references
See
“Generalized Method of Moments” and
“Panel Estimation” for discussion of the various GMM estimation techniques.
See
Equation::gmm for the equivalent equation object command.
Estimate a selection equation using the Heckman ML or 2-step method.
Syntax
heckit(options) response_eqn @ selection_eqn
The response equation should be the dependent variable followed by a list of regressors. The selection equation should be a binary dependent variable followed by a list of regressors.
Options
General Options
2step | Use the Heckman 2-step estimation method. Note that this option is incompatible with the maximum likelihood options below. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print the estimation results. |
ML Options
Note these options are not available if the "2step" option, above, is used.
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich methods)., |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
m=integer | Set maximum number of iterations. |
c=number | Set convergence criteria. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
s=number | Scale EViews’ starting values by number. |
r | Use Newton-Raphson optimizer. |
b | Use BHHH optimizer. |
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-1.txt
heckit ww c ax ax^2 we cit @ lfp c wa wa^2 faminc we (k618+kl6)>0
heckit(2step) ww c ax ax^2 we cit @ lfp c wa wa^2 faminc we (k618+kl6)>0
This example replicates the Heckman Selection example given in Greene (2008, page 888), which uses data from the Mroz (1987) study to estimate a selection model. The first line of this example downloads the data set, the second line creates an equation object and estimates it using the default maximum likelihood estimation method of Heckman Selection, which replicates the first pane of Table 24.3 in Greene. The third line estimates the same model, using the two-step approach, which replicates the second pane of Table 24.3.
Cross-references
Convert an entire Haver Analytics Database into an EViews database.
Syntax
hconvert haver_path db_name
You must have a Haver Analytics database installed on your computer to use this feature. You must also create an EViews database to store the converted Haver data before you use this command.
Be aware that this command may be very time-consuming.
Follow the command by a full path name to the Haver database and the name of an existing EViews database to store the Haver database. You can include a path name to the EViews database not in the default path.
Examples
dbcreate hdata
hconvert d:\haver\haver hdata
The first line creates a new (empty) database named HDATA in the default directory. The second line converts all the data in the Haver database and stores it in the HDATA database.
Cross-references
See
“EViews Databases” for a discussion of EViews and Haver databases.
Displays the documentation of an EViews command or procedure.
Syntax
help topic
Examples
help @cumsum
displays the documentation for the cumulative sum function.
help alpha.freq
displays the documentation for the alpha series freq procedure.
Fetch a series from a Haver Analytics database into a workfile.
hfetch reads one or more series from a Haver Analytics Database into the active workfile. You must have a Haver Analytics database installed on your computer to use this feature.
Syntax
hfetch(database_name) series_name
hfetch, if issued alone on a command line, will bring up a Haver dialog box which has fields for entering both the database name and the series names to be fetched. If you provide the database name (including the full path) in parentheses after the hfetch command, EViews will read the database and copy the series requested into the current workfile. It will also display information about the series on the status line. The database name is optional if a default database has been specified.
hfetch can read multiple series with a single command. List the series names, each separated by a space.
Examples
hfetch(c:\data\us1) pop gdp xyz
reads the series POP, GDP, and XYZ from the US1 database into the current active workfile, performing frequency conversions if necessary.
Cross-references
See also
“EViews Databases” for a discussion of EViews and Haver databases. Additional information on EViews frequency conversion is provided in
“Frequency Conversion”.
Histogram and descriptive statistics of a series.
The hist command computes descriptive statistics and displays a histogram for the series.
Syntax
hist(options) series_name
Options
Examples
hist lwage
Displays the histogram and descriptive statistics of LWAGE.
Cross-references
See
“Histogram and Stats” for a discussion of the descriptive statistics reported in the histogram view.
See
distplot for a more full-featured and customizable method of constructing histograms, and
Series::histfor the equivalent series object view command.
Display a Haver Analytics database series description.
hlabel reads the description of a series from a Haver Analytics Database and displays it on the status line at the bottom of the EViews window. Use this command to verify the contents of a Haver database series name.
You must have a Haver Analytics database installed on your computer to use this feature.
Syntax
hlabel(database_name) series_name
hlabel, if issued alone on a command line, will bring up a Haver dialog box which has fields for entering both the database name and the series names to be examined. If you provide the database name in parentheses after the hlabel command, EViews will read the database and find the key information about the series in question, such as the start date, end date, frequency, and description. This information is displayed in the status line at the bottom of the EViews window. Note that the database_name should refer to the full path to the Haver database but need not be specified if a default database has been specified in HAVERDIR.INI.
If several series names are placed on the line, hlabel will gather the information about each of them, but the information display may scroll by so fast that only the last will be visible.
Examples
hlabel(c:\data\us1) pop
displays the description of the series POP in the US1 database.
Cross-references
See
“EViews Databases” for a discussion of EViews and Haver databases.
See also
hfetch and
hconvert.
Smooth a series using the Hodrick-Prescott filter.
Syntax
hpf(options) series_name filtered_name [@ cycle_name]
Smoothing Options
The degree of smoothing may be specified as an option. You may specify the smoothing as a value, or using a power rule:
lambda=arg | Set smoothing parameter value to arg; a larger number results in greater smoothing. |
power=arg (default=2) | Set smoothing parameter value using the frequency power rule of Ravn and Uhlig (2002) (the number of periods per year divided by 4, raised to the power arg, and multiplied by 1600). Hodrick and Prescott recommend the value 2; Ravn and Uhlig recommend the value 4. |
If no smoothing option is specified, EViews will use the power rule with a value of 2.
Other Options
prompt | Force the dialog to appear from within a program. |
p | Print the graph of the smoothed series and the original series. |
Examples
hpf(lambda=1000) gdp gdp_hp
smooths the GDP series with a smoothing parameter “1000” and saves the smoothed series as GDP_HP.
Cross-references
See
Series::hpf for the equivalent series object command.
Imports data from a foreign file or a previously saved workfile into the current default workfile. The import command lets you perform four different types of data importing: dated reads, match-merge reads, sequential reads, and appends.
Dated imports can only be performed if the destination workfile is a dated workfile. You must specify the date structure of the source data as part of the import command. EViews will then match the date structure of the source with that of the destination, and perform frequency conversion if necessary.
Match-merge imports require both a source ID series and a destination ID series. EViews will read the source data into the destination workfile based upon matched values of the two ID series.
Sequential imports will read the source data into the destination workfile by matching the first observation of the source file to the first observation in the destination workfile's current sample, then the second observation of the source with the second observation in the destination's current sample, and so on.
Appended imports simply append the source data to the end of the destination workfile.
Syntax
The general form of the import command is:
import([type=], options) source_description import_specification [@smplsmpl_string] [genr_string] [@rename rename_string]
where the syntax for
import_specification depends on whether the read is a dated (
“Dated Imports”), match-merge (
“Match-Merge Import”), sequential (
“Sequential Read”), or appended import (
“Appended Read”).
• Source_description should contain a description of the file from which the data is to be imported. The specification of the description is usually just the path and file name of the file, however you can also specify more precise information. See
wfopen for more details on the specification of
source_description.
• The optional “type=” option may be used to specify a source type. For the most part, you should not need to specify a “type=” option as EViews will automatically determine the type from the filename. The following table summaries the various source formats and along with the corresponding “type=” keywords:
| |
Access | “access” |
Aremos-TSD | “a”, “aremos”, “tsd” |
Binary | “binary” |
dBASE | “dbase” |
Excel (through 2003) | “excel” |
Excel 2007 (xml) | “excelxml” |
EViews Workfile | --- |
Gauss Dataset | “gauss” |
GiveWin/PcGive | “g”, “give” |
HTML | “html” |
Lotus 1-2-3 | “lotus” |
ODBC Dsn File | “dsn” |
ODBC Query File | “msquery” |
ODBC Data Source | “odbc” |
MicroTSP Workfile | “dos”, “microtsp” |
MicroTSP Mac Workfile | “mac” |
RATS 4.x | “r”, “rats” |
RATS Portable / TROLL | “l”, “trl” |
SAS Program | “sasprog” |
SAS Transport | “sasxport” |
SPSS | “spss” |
SPSS Portable | “spssport” |
Stata | “stata” |
Text / ASCII | “text” |
TSP Portable | “t”, “tsp” |
• The optional @smpl keyword may be used to specify that data is only imported for the observations specified by smpl_string. By default, the import will use all of the observations in the workfile. If @smpl is included, but no smpl_string is included, then the current default workfile sample is used.
• The optional
@genr keyword may be used to generate a new series in the workfile as part of the import procedure.
genr_string may be any valid series creation expression, and can be an expression based upon one of the imported series. See
genr for information on valid expressions.
• The optional
@rename keyword may be used to rename some of the imported series where
rename_string contains pairs of old object names followed by the new names. See
rename for additional discussion.
In the remainder of this discussion, we examine each of the different import types in greater depth.
Dated Imports
The syntax for a dated import command is:
import([type=], options) source_description @freq frequency start_date [@smpl smpl_string] [genr_string] [@rename rename_string]
The
import_specification consists of the
@freq keyword followed by a
frequency argument specifying the frequency of the source data and a
start_date to specify the starting date of the source data. See
wfcreate for details on the forms that
frequency may take.
Basic Options
resize | Extend the destination workfile (if necessary) to include the entire range of the source data. |
link | Link the object to the source data so that the values can be refreshed at a later time. |
mode=arg (default=“o”) | Set the behavior for handling name conflicts when an imported series already exists in the destination workfile. arg can be “o” (Completely replace existing series with source series. Note that values outside of the range of the source data will be overwritten with NAs), “u” (Overwrite existing series only for values within the range of the source data. Destination values outside of the source range will be unchanged), “ms” (Overwrite existing series, unless source value is an NA, in which case keep destination values), “md” (Only overwrite NA values in destination series), “r” (rename any conflicts), or “p” (do not import any series which have a name conflict). |
page=page_name | Optional name for the page into which the data should be imported. |
tr=integer | Truncate long series names to integer characters. The default value of integer is 24. |
prompt | Force the dialog to appear from within a program. |
Frequency Conversion Options
The following options control the frequency conversion method when copying series and group objects into a workfile page and converting from low to high frequency:
c=arg | Low to high conversion methods: “r” or “repeata” (constant match average), “d” or “repeats” (constant match sum), “q” or “quada” (quadratic match average), “t” or “quads” (quadratic match sum), “linearf” (linear match first), “i” or “linearl” (linear match last), “cubicf” (cubic match first), “c” or “cubicl” (cubic match last), “pointf” (point first), “pointl” (point last). |
When importing data from a higher frequency source into a lower frequency destination:
c=arg | High to low conversion methods removing NAs: “a” (average of the nonmissing observations), “s” (sum of the nonmissing observations), “f” (first nonmissing observation), “l” (last nonmissing observation), “x” (maximum nonmissing observation), “m” (minimum nonmissing observation). High to low conversion methods propagating NAs: “an” or “na” (average, propagating missings), “sn” or “ns” (sum, propagating missings), “fn” or “nf” (first, propagating missings), “ln” or “nl” (last, propagating missings), “xn” or “nx” (maximum, propagating missings), “mn” or “nm” (minimum, propagating missings). |
Examples
import c:\temp\quarterly.xls @freq q 1990
will import the file QUARTERLY.XLS into the current default workfile. The source file has a quarterly frequency, starting at 1990.
import(c=s) c:\temp\quarterly.xls range="GDP_SHEET" @freq q 1990 @rename gdp_per_capita gdp
will import from same file, but instead will use the data on the Excel sheet called “GDP_SHEET”, and will rename the series GDP_PER_CAPITA to GDP. A frequency conversion method using the sum of the nonmissing observations is used rather than the default average.
import(mode=p) c:\temp\annual.txt @freq a 1990 @smpl 1994 1996
will import data from a text file called annual.txt, into the current default workfile. Any data in the text file that already exists in the destination workfile will be ignored, and for the remaining series, only the dates between 1994 and 1996 will be imported.
Additionally, specifying the “tr” option causes EViews 10 to truncate long series names to 24 characters (by default) instead of 300. This will let programs written for EViews 9 continue to work with EViews 10:
fetch(d=eia, link, tr) coal.average_employees.al-tot.a
The “tr” option can also be set to a number between 0 and 300:
fetch(d=eia, link, tr=20) coal.average_employees.al-tot.a
Match-Merge Import
Syntax
import(options) source_description @id id @destid destid [smpl_string] [genr_string] [@rename rename_string]
The import_specification consists of the @id keyword and at least one ID series in the source file, followed by the @destid keyword and at least one ID series in the destination workfile. The two sets of ID series should be compatible, in that they should contain a subset of identical values that provide information on how observations from the two files should be matched. If one of the ID series is a date series, you should surround it with the @date( ) keyword.
Options
resize | Extend the destination workfile (if necessary) to include the entire range of the source data. |
link | Link the object to the source data so that the values can be refreshed at a later time. |
mode=arg (default=“o”) | Set the behavior for handling name conflicts when an imported series already exists in the destination workfile. arg can be “o” (Completely replace existing series with source series. Note that values outside of the range of the source data will be overwritten with NAs), “u” (Overwrite existing series only for values within the range of the source data. Destination values outside of the source range will be unchanged), “ms” (Overwrite existing series, unless source value is an NA, in which case keep destination values), “md” (Only overwrite NA values in destination series), “r” (rename any conflicts), or “p” (do not import any series which have a name conflict). |
nacat | Treat “NA” values as a category when copying using general match merge operations. |
propnas | Propogate NAs / partial periods evaluate to NAs when converting. |
c=arg | Set the match merge contraction method. If you are importing a numeric source series by general match merge, the argument can be one of: “mean”, “med” (median), “max”, “min”, “sum”, “sumsq” (sum-of squares), “var” (variance), “sd” (standard deviation), “skew” (skewness), “kurt” (kurtosis), “quant” (quantile, used with “quant=” option), “obs” (number of observations), “nas” (number of NA values), “first” (first observation in group), “last” (last observation in group), unique” (single unique group value, if present), “none” (disallow contractions). If importing an alpha series, only the non-summary methods “max”, “min”, “obs”, “nas”, first”, “last”, “unique” and “none” are supported. For importing of numeric series, the default contraction method is “c=mean”; for copying of alpha series, the default is “c=unique”. |
page=page_name | Optional name for the page into which the data should be imported. |
prompt | Force the dialog to appear from within a program. |
Most of the conversion options should be self-explanatory. As for the others: “first” and “last” give the first and last non-missing observed for a given group ID; “obs” provides the number of non-missing values for a given group; “nas” reports the number of NAs in the group; “unique” will provide the value in the source series if it is the identical for all observations in the group, and will return NA otherwise; “none” will cause the import to fail if there are multiple observations in any group—this setting may be used if you wish to prohibit all contractions.
Examples
import(c=max, type=excel) c:\data\stateunemp.xls @id states @destid states
will import the file STATEUNEMP.XLS using the ID series STATES in both files as the match merge criteria. The maximum value is used as a contraction method. Note that although the “type=excel” option was used, it was not necessary since EViews will automatically detect the file type based on the file's extension (.xls).
import c:\data\stategdp.txt colhead=3 delim=comma @id states @date(year) @destid states @date
will import the file STATEGDP.TXT, specifying that there are three lines of column headers, and the delimiter for the text file is a comma. The series STATES is used as an ID series in both files, along with a date series (YEAR for the source file, and the default EViews date series, @DATE, for the destination workfile). Note that this type of import, with both a cross-section ID and a date ID is most commonly employed for importing data into panel workfiles.
import c:\data\cagdp.xls @id states @date(year) @destid states @date @genr states="CA"
will import the file CAGDP.XLS into the current workfile. In this particular case the source file is a time series file for a single state, California. Since the importing is being done into a panel workfile, the @genr keyword is used to generate a series containing the state identifier, CA, which is then used as the source ID.
Sequential Read
Syntax
import(options) source_description [@smplsmpl_string] [@genrgenr_string] [@rename rename_string]
No import_specification is required for a sequential read.
Options
resize | Extend the destination workfile (if necessary) to include the entire range of the source data. |
link | Link the object to the source data so that the values can be refreshed at a later time. |
mode=arg (default=“o”) | Set the behavior for handling name conflicts when an imported series already exists in the destination workfile. arg can be “o” (Completely replace existing series with source series. Note that values outside of the range of the source data will be overwritten with NAs), “u” (Overwrite existing series only for values within the range of the source data. Destination values outside of the source range will be unchanged), “ms” (Overwrite existing series, unless source value is an NA, in which case keep destination values), “md” (Only overwrite NA values in destination series), “r” (rename any conflicts), or “p” (do not import any series which have a name conflict). |
page=page_name | Optional name for the page into which the data should be imported. |
prompt | Force the dialog to appear from within a program. |
Examples
import(resize) sales.dta @smpl @all
will import the Stata file SALES.DTA into the current workfile, using the entire workfile range. If the sales.dta file contains more observations that the current workfile, the current workfile is resized to accommodate the extra rows of data.
Appended Read
Syntax
import(options) source_description @append[@genrgenr_string] [@rename rename_string]
The import_specification consists of the @append keyword. Note that the @smpl keyword is not supported for appended import.
Options
link | Link the object to the source data so that the values can be refreshed at a later time. |
mode=arg (default=“o”) | Set the behavior for handling name conflicts when an imported series already exists in the destination workfile. arg can be “o” (Completely replace existing series with source series. Note that values outside of the range of the source data will be overwritten with NAs), “r” (rename any conflicts), or “p” (do not import any series which have a name conflict). |
page=page_name | Optional name for the page into which the data should be imported. |
prompt | Force the dialog to appear from within a program. |
Examples
import(page=demand) demand.txt @append
will append the text file, DEMAND.TXT, to the bottom of the page “demand” in the current workfile.
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
Imports observation values stored inside one or more series in a second workfile page into the attribute fields of objects within the current workfile page.
Syntax
importattr(options) source_page [@keep keeplist @drop droplist]
The source page should contain one series whose values are the names of the objects into which the attributes should be imported, and one or more additional series containing the values of each attribute to be imported. By default, the procedure will assume there is an alpha series called NAME in the source page that contains the object names. The “name=” option can be used to specify that a different series in the source page contains the object names.
Values in the name column will always be converted into legal EViews names before matching to object names in the current workfile. (For example, any spaces will be replaced by underscores.)
Typically, the name of the series in the source workfile will be used as the attribute name inside the current workfile. However, if an object in the source workfile has a display name set, this value will be used for the attribute name instead. Display names must be used to specify attribute names if your attribute names contain spaces since spaces are not allowed within EViews object names.
For numeric series, attribute values will be imported using the formatting currently set for the series spreadsheet view of the source series.
By default, all series in the source page will be used as attribute values (except the series containing object names). To import only a subset of series as attributes, name patterns can be provided in the @keep and @drop arguments to restrict which series will be used.
importattr is most often useful when importing custom attributes from an external file. It is common for foreign data files, such as Excel files, to have one file (or sheet in Excel) containing the data, and a separate file (or sheet) containing the attributes, or meta-data, of each series. In such cases the pageload command can be used to read in the attribute file as a separate page in your workfile, and then importattr can be used to assign them to the data page.
Options
mode=arg | Specify how the procedure treats existing attribute values in the current workfile page. arg may be "o" or "overwrite" (clears all existing attribute values in the object before applying the imported attribute value), “u” or “update” (clears existing attribute values only for the attributes that are being imported), “m” or “merge” (keeps existing values if the imported value of the attribute is empty), “md” (keeps all existing non-empty values. only empty values will be replaced with the imported values). |
trim=string | Remove string from the start and end of the attribute name. |
name=arg | Specify the name of the alpha series in the source page containing the names of objects in the target page. |
Examples
importattr(name=objnames) attributes @keep attr1 attr2
Imports values from series ATTR1 and ATTR2 in the page “Attributes” into attributes “attr1” and “attr2” of objects in the current workfile. The series OBJNAMES in the page “Attributes” specifies which objects in the current workfile should have their attribute values updated.
importattr(name=objnames, trim=":") attributes @keep attr1 attr2
removes the “:” from the start and end of the attribute name, if present, since EViews adds that character in the label view.
Cross-references
See
“Workfile Details Display” and
“Label”for a discussion of the workfile details display and, for example, the series object label view.
Imports data from a foreign file into a matrix object in the current workfile.
Syntax
The general form of the importmat command is:
importmat([type=], options) source_description import_specification
• Source_description should contain a description of the file from which the data is to be imported. The specification of the description is usually just the path and file name of the file, however you can also specify more precise information. See
wfopen for more details on the specification of
source_description.
• The optional “type=” option may be used to specify a source type. For the most part, you should not need to specify a “type=” option as EViews will automatically determine the type from the filename. The following table summaries the various source formats and along with the corresponding “type=” keywords:
| |
Excel (through 2003) | “excel” |
Excel 2007 (xml) | “excelxml” |
HTML | “html” |
Text / ASCII | “text” |
Options
name=arg | Specify a name for the created matrix object. |
display=arg | Specify a display name for the created matrix object. |
page=page_name | Optional name for the page into which the matrix should be created. |
mode=o | Overwrite an existing object with the same name (only applicable if the name option is used). |
• import_specification can be used to provide additional information about the file to be read. The details of import_specification will depend upon the type of file being imported.
Excel Files
The syntax for reading Excel files is:
importmat(options) source_description [table_description] [variables_description]
The following table_description elements may be used when reading Excel data:
• “range = arg”, where arg is a range of cells to read from the Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the excel workbook to refer to a range or cell may be used to specify the cells to read.
• “byrow”, transpose the incoming data. This option allows you to read files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”. This option is rarely required.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int| all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Excel Examples
importmat "c:\data files\data.xls"
loads the active sheet of DATA.XLSX into a new untitled matrix object.
importmat(name=mymat) "c:\data files\data.xls" range="GDP data"
reads the data contained in the “GDP data” sheet of “Data.XLS” into the MYMAT object.
HTML Files
The syntax for reading HTML pages is:
importmat(options) source_description [table_description] [variables_description]
The following table_description elements may be used when reading an HTML file or page:
• “table = arg”, where arg specifies which HTML table to read in an HTML file/page containing multiple tables.
When specifying arg, you should remember that tables are named automatically following the pattern “Table01”, “Table02”, “Table03”, etc. If no table name is specified, the largest table found in the file will be chosen by default. Note that the table numbering may include trivial tables that are part of the HTML content of the file, but would not normally be considered as data tables by a person viewing the page.
• “skip = int”, where int is the number of rows to discard from the top of the HTML table.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”. This option is rarely used.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
HTML Examples
importmat "c:\data.html"
loads into a new untitled matrix object the data located in the HTML file “Data.HTML” located on the C:\ drive
importmat(type=html, name=forexmat) "http://www.tradingroom.com.au/apps/mkt/forex.ac" colhead=3
loads into a matrix object called FOREXMAT the data with the given URL located on the website site “http://www.tradingroom.com.au”. The column header is set to three rows.
Text and Binary Files
The syntax for reading text or binary files is:
importmat(options) source_description [table_description] [variables_description]
If a table_description is not provided, EViews will attempt to read the file as a free-format text file. The following table_description elements may be used when reading a text or binary file:
• “ftype = [ascii|binary]” specifies whether numbers and dates in the file are stored in a human readable text (ASCII), or machine readable (Binary) form.
• “rectype = [crlf|fixed|streamed]” describes the record structure of the file:
“crlf”, each row in the output table is formed using a fixed number of lines from the file (where lines are separated by carriage return/line feed sequences). This is the default setting.
“fixed”, each row in the output table is formed using a fixed number of characters from the file (specified in “reclen= arg”). This setting is typically used for files that contain no line breaks.
“streamed”, each row in the output table is formed by reading a fixed number of fields, skipping across lines if necessary. This option is typically used for files that contain line breaks, but where the line breaks are not relevant to how rows from the data should be formed.
• “reclines =int”, number of lines to use in forming each row when “rectype=crlf” (default is 1).
• “reclen=int”, number of bytes to use in forming each row when “rectype=fixed”.
• “recfields=int”, number of fields to use in forming each row when “rectype=streamed”.
• “skip=int”, number of lines (if rectype is “crlf”) or bytes (if rectype is not “crlf”) to discard from the top of the file.
• “comment=string“, where string is a double-quoted string, specifies one or more characters to treat as a comment indicator. When a comment indicator is found, everything on the line to the right of where the comment indicator starts is ignored.
• “emptylines=[keep|drop]”, specifies whether empty lines should be ignored (“drop”), or treated as valid lines (“keep”) containing missing values. The default is to ignore empty lines.
• “tabwidth=int”, specifies the number of characters between tab stops when tabs are being replaced by spaces (default=8). Note that tabs are automatically replaced by spaces whenever they are not being treated as a field delimiter.
• “fieldtype=[delim|fixed|streamed|undivided]”, specifies the structure of fields within a record:
“Delim”, fields are separated by one or more delimiter characters
“Fixed”, each field is a fixed number of characters
“Streamed”, fields are read from left to right, with each field starting immediately after the previous field ends.
“Undivided”, read entire record as a single series.
• “quotes=[single|double|both|none]”, specifies the character used for quoting fields, where “single” is the apostrophe, “double” is the double quote character, and “both” means that either single or double quotes are allowed (default is “both”). Characters contained within quotes are never treated as delimiters.
• “singlequote“, same as “quotes = single”.
• “delim=[comma|tab|space|dblspace|white|dblwhite]”, specifies the character(s) to treat as a delimiter. “White” means that either a tab or a space is a valid delimiter. You may also use the abbreviation “d=” in place of “delim=”.
• “custom="arg1"”, specifies custom delimiter characters in the double quoted string. Use the character “t” for tab, “s” for space and “a” for any character.
• “mult=[on|off]”, to treat multiple delimiters as one. Default value is “on” if “delim” is “space”, “dblspace”, “white”, or “dblwhite”, and “off” otherwise.
• “endian = [big|little]”, selects the endianness of numeric fields contained in binary files.
• “string = [nullterm|nullpad|spacepad]”, specifies how strings are stored in binary files. If “nullterm”, strings shorter than the field width are terminated with a single zero character. If “nullpad”, strings shorter than the field width are followed by extra zero characters up to the field width. If “spacepad”, strings shorter than the field width are followed by extra space characters up to the field width.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
• “lastcol”, include implied last column. For lines that end with a delimiter, this option adds an additional column. When importing a csv file, lines which have the delimiter as the last character (for example: “name, description, date”), EViews normally determines the line to have 3 columns. With the above option, EViews will determine the line to have 4 columns. Note this is not the same as a line containing “name, description, date”. In this case, EViews will always determine the line to have 3 columns regardless if the option is set.
A central component of the table_description element is the format statement. You may specify the data format using the following table descriptors:
• Fortran Format:
fformat=([n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)
where Type specifies the underlying data type, and may be one of the following,
I - integer
F - fixed precision
E - scientific
A - alphanumeric
X - skip
and n1, n2, ... are the number of times to read using the descriptor (default=1). More complicated Fortran compatible variations on this format are possible.
• Column Range Format:
rformat="[n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)"
where optional type is “$” for string or “#” for number, and n1, n2, n3, n4, etc. are the range of columns containing the data.
• C printf/scanf Format:
cformat="fmt"
where fmt follows standard C language (printf/scanf) format rules.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”. This option is rarely used.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Text and Binary File Examples (.txt, .csv, etc.)
importmat c:\data.csv skip=5
reads “Data.CSV” into a new unnamed matrix object, skipping the first 5 rows.
importtbl(type=text, name=matrix01) c:\date.txt delim=comma
loads the comma delimited data DATE.TXT into the MATRIX01 matrix object.
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
Imports data from a foreign file into a table object in the current workfile.
Syntax
The general form of the importtbl command is:
importtbl([type=], options) source_description import_specification
• Source_description should contain a description of the file from which the data is to be imported. The specification of the description is usually just the path and file name of the file, however you can also specify more precise information. See
wfopen for more details on the specification of
source_description.
• The optional “type=” option may be used to specify a source type. For the most part, you should not need to specify a “type=” option as EViews will automatically determine the type from the filename. The following table summaries the various source formats and along with the corresponding “type=” keywords:
| |
Excel (through 2003) | “excel” |
Excel 2007 (xml) | “excelxml” |
HTML | “html” |
Text / ASCII | “text” |
Options
name=arg | Specify a name for the created table object. |
display=arg | Specify a display name for the created table object. |
page=page_name | Optional name for the page into which the table should be created. |
mode=o | Overwrite an existing object with the same name (only applicable if the name option is used). |
• import_specification can be used to provide additional information about the file to be read. The details of import_specification will depend upon the type of file being imported.
Excel Files
The syntax for reading Excel files is:
importtbl(options) source_description [table_description] [variables_description]
The following table_description elements may be used when reading Excel data:
• “range = arg”, where arg is a range of cells to read from the Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the excel workbook to refer to a range or cell may be used to specify the cells to read.
• “byrow”, transpose the incoming data. This option allows you to read files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”. This option is rarely required.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int| all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Excel Examples
importtbl "c:\data files\data.xls"
loads the active sheet of DATA.XLSX into a new untitled table object.
importtbl(name=mytbl) "c:\data files\data.xls" range="GDP data"
reads the data contained in the “GDP data” sheet of “Data.XLS” into the MYTBL object.
HTML Files
The syntax for reading HTML pages is:
importtbl(options) source_description [table_description] [variables_description]
The following table_description elements may be used when reading an HTML file or page:
• “table = arg”, where arg specifies which HTML table to read in an HTML file/page containing multiple tables.
When specifying arg, you should remember that tables are named automatically following the pattern “Table01”, “Table02”, “Table03”, etc. If no table name is specified, the largest table found in the file will be chosen by default. Note that the table numbering may include trivial tables that are part of the HTML content of the file, but would not normally be considered as data tables by a person viewing the page.
• “skip = int”, where int is the number of rows to discard from the top of the HTML table.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”. This option is rarely used.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
HTML Examples
importtbl "c:\data.html"
loads into a new untitled table object the data located in the HTML file “Data.HTML” located on the C:\ drive
importtbl(type=html, name=forextbl) "http://www.tradingroom.com.au/apps/mkt/forex.ac" colhead=3
loads into a table object called FOREXTBL the data with the given URL located on the website site “http://www.tradingroom.com.au”. The column header is set to three rows.
Text and Binary Files
The syntax for reading text or binary files is:
importtbl(options) source_description [table_description] [variables_description]
If a table_description is not provided, EViews will attempt to read the file as a free-format text file. The following table_description elements may be used when reading a text or binary file:
• “ftype = [ascii|binary]” specifies whether numbers and dates in the file are stored in a human readable text (ASCII), or machine readable (Binary) form.
• “rectype = [crlf|fixed|streamed]” describes the record structure of the file:
“crlf”, each row in the output table is formed using a fixed number of lines from the file (where lines are separated by carriage return/line feed sequences). This is the default setting.
“fixed”, each row in the output table is formed using a fixed number of characters from the file (specified in “reclen= arg”). This setting is typically used for files that contain no line breaks.
“streamed”, each row in the output table is formed by reading a fixed number of fields, skipping across lines if necessary. This option is typically used for files that contain line breaks, but where the line breaks are not relevant to how rows from the data should be formed.
• “reclines =int”, number of lines to use in forming each row when “rectype=crlf” (default is 1).
• “reclen=int”, number of bytes to use in forming each row when “rectype=fixed”.
• “recfields=int”, number of fields to use in forming each row when “rectype=streamed”.
• “skip=int”, number of lines (if rectype is “crlf”) or bytes (if rectype is not “crlf”) to discard from the top of the file.
• “comment=string“, where string is a double-quoted string, specifies one or more characters to treat as a comment indicator. When a comment indicator is found, everything on the line to the right of where the comment indicator starts is ignored.
• “emptylines=[keep|drop]”, specifies whether empty lines should be ignored (“drop”), or treated as valid lines (“keep”) containing missing values. The default is to ignore empty lines.
• “tabwidth=int”, specifies the number of characters between tab stops when tabs are being replaced by spaces (default=8). Note that tabs are automatically replaced by spaces whenever they are not being treated as a field delimiter.
• “fieldtype=[delim|fixed|streamed|undivided]”, specifies the structure of fields within a record:
“Delim”, fields are separated by one or more delimiter characters
“Fixed”, each field is a fixed number of characters
“Streamed”, fields are read from left to right, with each field starting immediately after the previous field ends.
“Undivided”, read entire record as a single series.
• “quotes=[single|double|both|none]”, specifies the character used for quoting fields, where “single” is the apostrophe, “double” is the double quote character, and “both” means that either single or double quotes are allowed (default is “both”). Characters contained within quotes are never treated as delimiters.
• “singlequote“, same as “quotes = single”.
• “delim=[comma|tab|space|dblspace|white|dblwhite]”, specifies the character(s) to treat as a delimiter. “White” means that either a tab or a space is a valid delimiter. You may also use the abbreviation “d=” in place of “delim=”.
• “custom="arg1"”, specifies custom delimiter characters in the double quoted string. Use the character “t” for tab, “s” for space and “a” for any character.
• “mult=[on|off]”, to treat multiple delimiters as one. Default value is “on” if “delim” is “space”, “dblspace”, “white”, or “dblwhite”, and “off” otherwise.
• “endian = [big|little]”, selects the endianness of numeric fields contained in binary files.
• “string = [nullterm|nullpad|spacepad]”, specifies how strings are stored in binary files. If “nullterm”, strings shorter than the field width are terminated with a single zero character. If “nullpad”, strings shorter than the field width are followed by extra zero characters up to the field width. If “spacepad”, strings shorter than the field width are followed by extra space characters up to the field width.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
A central component of the table_description element is the format statement. You may specify the data format using the following table descriptors:
• Fortran Format:
fformat=([n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)
where Type specifies the underlying data type, and may be one of the following,
I - integer
F - fixed precision
E - scientific
A - alphanumeric
X - skip
and n1, n2, ... are the number of times to read using the descriptor (default=1). More complicated Fortran compatible variations on this format are possible.
• Column Range Format:
rformat="[n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)"
where optional type is “$” for string or “#” for number, and n1, n2, n3, n4, etc. are the range of columns containing the data.
• C printf/scanf Format:
cformat="fmt"
where fmt follows standard C language (printf/scanf) format rules.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”. This option is rarely used.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Text and Binary File Examples (.txt, .csv, etc.)
importtbl c:\data.csv skip=5
reads “Data.CSV” into a new unnamed table object, skipping the first 5 rows.
importtbl(type=text, name=table01) c:\date.txt delim=comma
loads the comma delimited data DATE.TXT into the TABLE01 table object.
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
Limited Information Maximum Likelihood and K-class Estimation.
Syntax
liml(options) y c x1 [x2 x3 ...] @ z1 [z2 z3 ...]
liml(options) specification @ z1 [z2 z3 ...]
To use the liml command, list the dependent variable first, followed by the regressors, then any AR or MA error specifications, then an “@”-sign, and finally, a list of exogenous instruments.
You may estimate nonlinear equations or equations specified with formulas by first providing a specification, then listing the instrumental variables after an “@”-sign. There must be at least as many instrumental variables as there are independent variables. All exogenous variables included in the regressor list should also be included in the instrument list. A constant is included in the list of instrumental variables, unless the noconst option is specified.
Options
noconst | Do not include a constant in the instrumental list. Without this option, a constant will always be included as an instrument, even if not specified explicitly. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
kclass=number | Set the value of 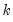 in the K‑class estimator. If omitted, LIML is performed, and 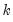 is calculated as part of the estimation procedure. |
se = arg (default=“iv”) | Set the standard-error calculation type: IV based (“se=iv”), K-Class based (“se=kclass”), Bekker (“se=bekk”), or Hansen, Hausman, and Newey (“se=hhn”). |
m=integer | Set maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. Available only for legacy estimation (“optmeth=legacy”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
liml gdp c cpi inc @ lw lw(-1)
calculates a LIML estimation of GDP on a constant, CPI, and INC, using a constant, LW, and LW(-1) as instruments.
Cross-references
See
Equation::liml for the equivalent equation object method.
Load a workfile.
Provided for backward compatibility. Same as
wfopen.
Clears the log window corresponding to the program.
Use this command any time from within a program to clear the program log.
Syntax
logclear(options)
Options
name=arg | Name of the message log. |
filename=arg | Name of the message log file to store the messages. |
Examples
To clear the contents of the message log FIRSTRUN:
logclear(name=”firstrun”)
To clear the contents of the message log FIRSTRUN and delete the file C:\MYFILE.TXT:
logclear(name=”firstrun”, filename=”c:\myfile.txt”)
Cross-references
Closes one or more or all message log windows.
Use this command any time from the command line or from within a program to close the log windows.
Syntax
logclose logwindow_list
where logwindow_list is a list of log windows to close: “@all” will close all log windows, while “arg1 arg2 arg3” will close the log windows ARG1, ARG2, and ARG3.
Examples
To close all log windows:
logclose @all
To close the log windows FIRSTRUN and SECONDRUN:
logclose firstrun secondrun
Cross-references
Sends result of the command to a log window.
Syntax
logeval command
Examples
logeval @pagelist
Sends to a log window a string containing the names of all the pages in the current active workfile.
logeval @abs(3)
Sends to a log window the absolute value of 3.
Cross-references
Estimate binary models with logistic errors.
Provide for backward compatibility. Equivalent to issuing the command, binary with the option “(d=l)”.
Set the log settings of specified message types.
Activates or deactivates the logging of specified message types if message types are set to be program controlled.
Controls the output of the messages to a log window with a specified name and/or to a file on disk.
Syntax
logmode(options) msgtype_list
where msgtype_list is a list of the message types to update.
Options
name=arg | Name of the message log and message log window. Default name is: PROGRAM_NAME. |
filename=arg | Name of the message log file on disk to store the messages. |
autosave=on/off | Turns on/off automatic saving of the message logs on disk. |
clearfile | Deletes the message log file (specified by filename option). |
clear | Clears the log window. |
float | Makes the log window floating. |
timestamp, ts | Adds a timestamp in front of the message. |
Message type options
all/-all | [Show/Do not show] all messages. |
error/-error, e/-e | [Show/Do not show] error messages. |
logmsg /-logmsg, l/ -l | [Show/Do not show] logmsg messages. |
program/-program, p/-p | [Show/Do not show] program lines. |
statusline/-statusline, s / -s | [Show/Do not show] status line messages. |
hideprogline/-hideprogline | [Hide/Do not hide] the program line when reporting errors encountered during execution. |
addin/-addin | [Show/Do not show] messages generally appropriate for addin error reporting. addin is equivalent to the command and program mode (
“Program Modes”) statements: logmode hideprogline -error mode quiet |
debug | Show messages generally appropriate for debugging of programs. Equivalent to the command: logmode -hideprogline error |
Note that using logmode with debug will override all subsequent (in either the current program or any program run using exec or run), logmode with hideprog and -error specifications. In particular, debug will override subsequent logmode addin statements.
Examples
logmode p
turns on logging and directs the output to the window PROGRAM_NAME. Note that by default all message types are initially turned off.
logmode error -p
Building on the first command, this activates the logging of errors and deactivates the logging of program lines. The error messages will be displayed on a window with name PROGRAM_NAME.
logmode -all s
turns off logging of all types, with the exception of status line messages. The status line messages will be displayed on a window with name PROGRAM_NAME. Note the order of message types is important. For example,
logmode p -all
will initially activate the logging of program lines, but following p with -all will deactivate program lines as well as any other messages that have been previously activated. The program lines will be displayed in a window with name PROGRAM_NAME
logmode(name="firstrun") l
turns on logging of logmsg messages and outputs the messages on a window with name FIRSTRUN.
logmode(name="") l
Building on the previous command, this will no longer output the messages on the window with name FIRSTRUN, but instead on a window with name PROGRAM_NAME.
logmode(name="secondrun", filename="c:\myfile.txt", autosave=on)
turns on logging, sends the output to the window SECONDRUN, and writes the messages to a file with name myFile.txt.
logmode(name="thirdrun", clear, ts, float, autosave=off)
directs messages to a window with name THIRDRUN, clears the window, adds a timestamp to all messages, makes the window floating, and turns off the autosave.
logmode(name="fourthrun", autosave=on )
turns on logging, sends messages to a window with name fourthrun, and writes messages to a file with name program_name.txt
Cross-references
Adds a line of text to the program log.
Syntax
logmsg(options) text
Options
name=arg | Name of the message log where messages will be added. |
filename=arg | Name of the message log where messages will be stored. |
Example
logmsg About to iterate through list of states
appends the text “About to iterate through list of states” to the program log with name PROGRAM_NAME.
logmsg(name="firstrun") Exiting list iteration
appends the text "Exiting list iteration" on the program log with name FIRSTRUN.
logmsg(filename="c:\myfile.txt") Restarting list iteration
appends the text "Restarting list iteration" on the program log with name FIRSTRUN. It also stores the message in the file c:\myfile.txt.
Cross-references
Saves the program log to a text file.
Syntax
logsave(options) destination
where destination is the location and filename for saving the log file (for types text and rtf) or the text object name (for type textobj)
Options
type=arg | Type TEXT to save the log as a TXT file (default). Type RTF to save the log as an RTF file (to preserve syntax coloring). Type TEXTOBJ to save the log as a TEXTOBJECT in the current workfile. |
name=arg | Name of the message log, the contents of which will be written in the log file. |
append | Append to the file, if the file exists. Default behavior clears the contents of the file. (Not supported for type=rtf). |
Example
logsave c:\EViews\myprog.text
saves the contents of the program log to the text file MYPROG, in the “C:\EViews” directory.
logsave(name="firstrun", type=rtf) c:\EViews\myprog.rtf
saves the contents of the program log FIRSTRUN to the rtf file MYPROG, in the "C:\EViews" directory.
logsave(name="secondrun", type=textobj) mytextobj
saves the contents of the program log SECONDRUN to a text object MYTEXTOBJ in the current workfile.
logsave(name="thirdrun", type=text, append) c:\EViews\myprog.txt
saves the contents of the program log THIRDRUN to the text file MYPROG, in the "C:\EViews" directory. The output will be appended at the end of the file.
Cross-references
When the current workfile has a panel structure, ls also estimates cross-section weighed least squares, feasible GLS, and fixed and random effects models.
Syntax
ls(options) y x1 [x2 x3 ...]
ls(options) specification
For linear specifications, list the dependent variable first, followed by a list of the independent variables. Use a “C” if you wish to include a constant or intercept term; unlike some programs, EViews does not automatically include a constant in the regression. You may add AR, MA, SAR, and SMA error specifications, a D fractional differencing term, and PDL specifications for polynomial distributed lags. If you include lagged variables, EViews will adjust the sample automatically, if necessary.
Both dependent and independent variables may be created from existing series using standard EViews functions and transformations. EViews treats the equation as linear in each of the variables and assigns coefficients C(1), C(2), and so forth to each variable in the list.
Linear or nonlinear single equations may also be specified by explicit equation. You should specify the equation as a formula. The parameters to be estimated should be included explicitly: “C(1)”, “C(2)”, and so forth (assuming that you wish to use the default coefficient vector “C”). You may also declare an alternative coefficient vector using coef and use these coefficients in your expressions.
Options
Non-Panel LS Options
indicator | Include indicator saturation detection as part of estimation routine. |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
z | Turn off backcasting in ARMA models where “arma=cls”. |
optmethod = arg | Optimization method for nonlinear least squares and ARMA: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “kohn” (Kohn-Ansley for ARMA estimated by ML or GLS), or “legacy” (EViews legacy for nonlinear least squares and ARMA estimated by CLS). Gauss-Newton is the default method. |
optstep = arg | Step method for nonlinear least squares and ARMA: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method available for nonlinear least squares or ARMA estimated by CLS), “hac” (Newey-West HAC, available for nonlinear least squares or ARMA estimated by CLS).. |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 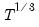 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
arma=arg | ARMA estimation method: “ml” (maximum likelihood); “gls” (generalized least squares), “cls” (conditional least squares). Not applicable to ARFIMA models which always estimate using maximum likelihood. |
armastart=arg | ARMA coefficient starting values: “auto” (automatic) “fixed” (legacy EViews fixed); “random” (random draw); “user” (user-specified). Applicable when “arma=ml” or “arma=gls”. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR or MA terms when “arma=cls” (see also
param ). |
s=number | Determine starting values for equations specified by list with AR or MA terms when “arma=cls”. Specify a number between zero and one representing the fraction of preliminary least squares estimates computed without AR or MA terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR and MA terms which are set to EViews determined default values. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. Available only for legacy estimation (“optmeth=legacy”). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method available for nonlinear least squares or ARMA estimated by CLS), “hac” (Newey-West HAC, available for nonlinear least squares or ARMA estimated by CLS)., “hc” (extended heteroskedasticity consistent), “hcuser” (user-specified heteroskedasticity), “cr” (cluster robust). The extended “hc” methods are only available for linear specifications. |
hctype=arg (default “hc2”) | Extended heteroskedasticity consistent method: “hc0” (no d.f. adjustment), “hc1” (d.f. adjusted), “hc2”, “hc3”, “hc4”, “hc4m”, “hc5”, when “cov=hc”. |
userwt=arg | Name of series containing user-diagonal weights (if “cov=hcuser”) |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), “hc2”, “hc3”, “hc4”, “hc4m”, “hc5”, when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
k=arg (default = 0.7) | Parameter for “cov=hc, hctype=hc5” or “cov=cr, crtype=cr5”. |
k1=arg (default = 1.0) | Parameter for “cov=hc, hctype=hc4m” or “cov=cr, crtype=cr4m”. |
k2=arg (default = 1.5) | Parameter for “cov=hc, hctype=hc4m” or “cov=cr, crtype=cr4m”. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Note: not all options are available for all equation methods.
Non-Panel Indicator Saturation Options
For use if “indicator” option is specified.
noiis | Do not search for impulse terms. |
sis | Search for step-shift terms. |
trend | Search for trend terms. |
pval=number (default = 0.05) | Set the terminal condition p-value used to determine the stopping point of each search path |
nolm | Do not perform AR LM diagnostic test. |
arpval=number (default = 0.025) | Set p-value used in AR LM diagnostic test. |
arlags=int (default = 1) | Set number of lags used in AR LM diagnostic test. |
noarch | Do not perform ARCH LM diagnostic test. |
archpval=number (default = 0.025) | Set p-value used in ARCH LM diagnostic test. |
archlags=int (default = 1) | Set number of lags used in ARCH LM diagnostic test. |
nojb | Do not perform Jarque-Bera normality diagnostic test. |
jbpval=number (default = 0.025) | Set p-value used in Jarque-Bera normality diagnostic test. |
nopet | Do not perform Parsimonious Encompassing diagnostic test. |
petpval=number (default = 0.025) | Set p-value used in Parsimonious Encompassing diagnostic test. |
nogum | Do not include the general model as a candidate for model selection. |
noempty | Do not include the empty model as a candidate for model selection. |
ic =arg | Set the information criterion used in model selection: “AIC” (Akaike information criteria, default), “BIC” (Schwarz information criteria), “HQ” (Hannan-Quin criteria). |
blocks=int | Override the EViews’ determination of the number of blocks in which to split the estimation sample. |
Panel LS Options
cx=arg | Cross-section effects: (default) none, fixed effects (“cx=f”), random effects (“cx=r”). |
per=arg | Period effects: (default) none, fixed effects (“per=f”), random effects (“per=r”). |
wgt=arg | GLS weighting: (default) none, cross-section system weights (“wgt=cxsur”), period system weights (“wgt=persur”), cross-section diagonal weighs (“wgt=cxdiag”), period diagonal weights (“wgt=perdiag”). |
cov=arg | Coefficient covariance method: (default) ordinary, White cross-section system (period clustering) robust (“cov=cxwhite” or “cov=percluster”), White period system (cross-section clustering) robust (“cov=perwhite” or “cov=cxcluster”), White heteroskedasticity robust (“cov=stackedwhite”), White two-way cluster robust (cov=bothcluster”), Cross-section system robust/PCSE (“cov=cxsur”), Period system robust/PCSE (“cov=persur”), Cross-section heteroskedasticity robust/PCSE (“cov=cxdiag”), Period heteroskedasticity robust/PCSE (“cov=perdiag”). |
keepwgts | Keep full set of GLS weights used in estimation with object, if applicable (by default, only small memory weights are saved). |
rancalc=arg (default=“sa”) | Random component method: Swamy-Arora (“rancalc=sa”), Wansbeek-Kapteyn (“rancalc=wk”), Wallace-Hussain (“rancalc=wh”). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
iter=arg (default= “onec”) | Iteration control for GLS specifications: perform one weight iteration, then iterate coefficients to convergence (“iter=onec”), iterate weights and coefficients simultaneously to convergence (“iter=sim”), iterate weights and coefficients sequentially to convergence (“iter=seq”), perform one weight iteration, then one coefficient step (“iter=oneb”). Note that random effects models currently do not permit weight iteration to convergence. |
unbalsur | Compute SUR factorization in unbalanced data using the subset of available observations for a cluster. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR terms (see also
param ). |
s=number | Determine starting values for equations specified by list with AR terms. Specify a number between zero and one representing the fraction of preliminary least squares estimates computed without AR terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR terms which are instead set to EViews determined default values. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
ls m1 c uemp inf(0 to -4) @trend(1960:1)
estimates a linear regression of M1 on a constant, UEMP, INF (from current up to four lags), and a linear trend.
ls(z) d(tbill) c inf @seas(1) @seas(1)*inf ma(2)
regresses the first difference of TBILL on a constant, INF, a seasonal dummy, and an interaction of the dummy and INF, with an MA(2) error. The “z” option turns off backcasting.
coef(2) beta
param beta(1) .2 beta(2) .5 c(1) 0.1
ls(cov=white) q = beta(1)+beta(2)*(l^c(1) + k^(1-c(1)))
estimates the nonlinear regression starting from the specified initial values. The “cov=white” option reports heteroskedasticity consistent standard errors.
ls r = c(1)+c(2)*r(-1)+div(-1)^c(3)
estimates an UNTITLED nonlinear equation.
ls(cx=f, per=f) n w k ys c
estimates an UNTITLED equation in a panel workfile using both cross-section and period fixed effects.
ls(cx=f, wgt=cxdiag) n w k ys c
estimates an UNTITLED equation in a panel workfile with cross-section weights and fixed effects.
Cross-references
“Basic Regression Analysis” and
“Additional Regression Tools” discuss the various regression methods in greater depth.
“Special Expression Reference” describes special terms that may be used in
ls specifications.
See
“Panel Estimation” for a discussion of panel equation estimation.
See
Equation::ls for the equivalent equation object method command.
Place submatrix in matrix.
Place matrix object at a specified location in a matrix.
Syntax
matplace(m1, m2[, n1, n2])
Places the matrix object m2 into m1 at row n1 and column n2. The sizes of the two matrices do not matter, as long as m1 is large enough to contain all of m2 with the upper left cell of m2 placed at row n1 and column n2.
Examples
matrix(100,5) m1
matrix(100,2) m2
matplace(m1, m2, 1, 1)
Cross-references
Estimates an equation using Mixed Data Sampling (MIDAS) regression.
MIDAS regression is an estimation technique which allows for data sampled at different frequencies to be used in the same regression.
Syntax
midas(options) y x1 [x2 x3 ...] @ z1page\z1 [z2page\z2 ...]
where y, x1, etc., are the dependent and explanatory variables in the current page frequency, and z1page\z1 and z2page\z2 are the high frequency variable page\series specification.
You may not include ARMA terms in a MIDAS regression.
Options
General options
midwgt=arg | MIDAS weight method: step function(“step”), normalized exponential Almon (“expalmon”), normalized beta function (“beta”), U-MIDAS (“umidas”), Auto-search/GETS (“autogets”) or the default Almon/PDL weighting (“almon”). |
lag=arg | Method for specifying the number of lags of the high frequency regressor to use: lag selection (“auto”), fixed (“fixed”). The default is “lag=fixed”. |
maxlag=arg | Maximum number of lags of the high frequency regressor to use when using lag selection. For use when “lag=auto”. The default value is 4. |
fixedlag=arg | Fixed number of lags of the high frequency regressor to use. For use when “lags=fixed”. The default value is 4. |
steps=integer | Stepsize (number of high frequency periods to group). For use when “midwgt=step”. |
polynomial=integer | Polynomial degree. For use when Almon/PDL weighting is employed. |
beta=arg | Beta function restriction: none (“none”), trend coefficient equals 1 (“trend”), endpoints coefficient equals 0 (“endpoint”), both trend and endpoints restriction (“both”). For use when “midwgt=beta”. The default is “beta=none”. |
optmethod = arg | Optimization method for nonlinear estimation: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “hybrid” (initial BHHH followed by BFGS). Hybrid is the default method. |
optstep = arg | Step method for nonlinear estimation: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method for nonlinear models: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich). |
covinfo = arg | Information matrix method for nonlinear models: “opg” (OPG); “hessian” (observed Hessian). |
freq = key | Set the frequency conversion method. Key can be “first” (the higher frequency data are used from the first observation in the lower frequency period), “last” (default, the higher frequency data are used from the last observation in the lower frequency), or “match” (a specific date matching series from each page is used). |
freqsrc = arg | Set the source date matching series. Only applies if freq=match is used. |
freqdest = arg | Set the destination date matching series. Only applies if freq=match is used. |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values in nonlinear estimation (see also
param ). |
s=number | Determine starting values for nonlinear estimation.. Specify a number between zero and one representing the fraction of preliminary EViews chosen values. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Auto-search/GETS options
pval=number (default = 0.05) | Set the terminal condition p-value used to determine the stopping point of each search path |
nolm | Do not perform AR LM diagnostic test. |
arpval=number (default = 0.025) | Set p-value used in AR LM diagnostic test. |
arlags=int (default = 1) | Set number of lags used in AR LM diagnostic test. |
noarch | Do not perform ARCH LM diagnostic test. |
archpval=number (default = 0.025) | Set p-value used in ARCH LM diagnostic test. |
archlags=int (default = 1) | Set number of lags used in ARCH LM diagnostic test. |
nojb | Do not perform Jarque-Bera normality diagnostic test. |
jbpval=number (default = 0.025) | Set p-value used in Jarque-Bera normality diagnostic test. |
nopet | Do not perform Parsimonious Encompassing diagnostic test. |
petpval=number (default = 0.025) | Set p-value used in Parsimonious Encompassing diagnostic test. |
nogum | Do not include the general model as a candidate for model selection. |
noempty | Do not include the empty model as a candidate for model selection. |
ic =arg | Set the information criterion used in model selection: “AIC” (Akaike information criteria, default), “BIC” (Schwarz information criteria), “HQ” (Hannan-Quin criteria). |
blocks=int | Override the EViews’ determination of the number of blocks in which to split the estimation sample. |
Examples
midas(fixedlag=9, midwgt=beta, beta=endpoint) realgdp c realgdp(-1) @ monthlypage\emp(-5)
estimates a MIDAS beta weight specification using the low frequency dependent variable REALGDP and regressors C and REALGDP(-1), and 9 beta weighted lags of EMP(-5) from the “monthlypage” workfile page. The beta weight function places zero restrictions on the endpoint coefficient.
midas(maxlag=12, lag=auto) realgdp c realgdp(-1) @ monthlypage\emp(-5)
estimates the same equation using PDL/Almon weights. The number of lags is chosen automatically with a maximum of 12 lags.
Cross-references
“Midas Regression” discusses the specification and estimation of MIDAS regression models in EViews.
Convert matrix object to a series or group.
atrix-TO-eries object: convert the data in a matrix object to a series, alpha, or group.
Syntax
mtos(vector, series[, sample])
mtos(svector, alpha[, sample])
mtos(matrix, group[, sample, prefix])
mtos(matrix, group[, prefix])
The number of observations in the sample must match the row size of the matrix to be converted. If no sample is provided, the matrix is written into the series using the current workfile sample.
For the matrix forms of the command, the prefix parameter is a string. If the target group object does not exist, the group is created and populated with series named <prefix>1, <prefix>2, etc. If the prefix is omitted, the default prefix is “SER”.
Examples
mtos(mom,gr1)
converts the first column of the matrix MOM to the first series in the group GR1, the second column of MOM to the second series in GR1, and so on. The current workfile sample length must match the row length of the matrix MOM. If GR1 is an existing group object, the number of series in GR1 must match the number of columns of MOM. If a group object named GR1 does not exist, EViews creates GR1 with the first series named SER1, the second series named SER2, and so on.
series col1
series col2
group g1 col1 col2
sample s1 1951 1990
mtos(m1,g1,s1)
The first two lines declare series objects, the third line declares a group object, the fourth line declares a sample object, and the fifth line converts the columns of the matrix
M1 to series in group
G1 using sample
S1. This command will generate an error if
M1 is not a
matrix.
Cross-references
See
“Matrix Language”for further discussion and examples of the use of matrices.
See also
stom,
stomna, and
@convert.
Generate random normal draws.
The nrnd command fills series, vector, and matrix objects with (pseudo) random values drawn from a standard normal distribution. When used with a series, the nrnd command ignores the current sample and fills the entire object.
Syntax
nrnd(object_name)
Fill object_name with normal random numbers.
Examples
matrix(10, 3) m1
nrnd(m1)
Cross-references
For random generator functions, see
“Statistical Distributions” and in particular,
@mrnd,
@mnrnd, and
@rmvnorm.
For related commands, see
rnd,
rndint, and
rmvnorm. See also
rndseed.
Opens a program file, or text (ASCII) file.
This command should be used to open program files or text (ASCII) files for editing.
You may also use the command to open workfiles or databases. This use of the
open command for this purposes is provided for backward compatibility. We recommend instead that you use the new commands
wfopen and
pageload to open a workfile, and
dbopen to open databases.
Syntax
open(options) [path\]file_name
You should provide the name of the file to be opened including the extension (and optionally a path), or the file name without an extension but with an option identifying its type. Specified types always take precedence over automatic type identification. If a path is not provided, EViews will look for the file in the default directory.
Files with the “.PRG” extension will be opened as program files, unless otherwise specified. Files with the “.TXT” extension will be opened as text files, unless otherwise specified.
For backward compatibility, files with extensions that are recognized as database files are opened as EViews databases, unless an explicit type is specified. Similarly, files with the extensions “.WF” and “.WF1”, and foreign files with recognized extensions will be opened as workfiles, unless otherwise specified.
All other files will be read as text files.
Options
p | Open file as program file. |
t | Open file as text file. |
type=arg (“prg” or “txt”) | Specify text or program file type using keywords. |
Examples
open finfile.txt
opens a text file named “Finfile.TXT” in the default directory.
open "c:\program files\my files\test1.prg"
opens a program file named “Test1.PRG” from the specified directory.
open a:\mymemo.tex
opens a text file named “Mymemo.TEX” from the A: drive.
Cross-references
See
wfopen and
pageload for commands to open files as workfiles or workfile pages, and
dbopen for opening database files.
Find the solution to a user-defined optimization problem.
The optimize command calls the EViews optimizer to find the optimal control values for a subroutine defined elsewhere in the program file or files.
Syntax
optimize(options) subroutine_name(arguments)
You should follow the optimize command keyword with options and subroutine_name, the name of a defined subroutine in your program (or included programs).
The subroutine must contain at least two arguments.
By default EViews will interpret the first argument as the output of the subroutine and will use it as the value to optimize. If the objective contains more than one value, as in a series or vector, EViews will optimize the sum of the values. The second argument is, by default, used to define the control parameters for which EViews will find the optimal values.
Since the objective and controls are defined by a standard EViews subroutine, the arguments of the subroutine may correspond to numbers, strings, and various EViews objects such as series, vectors, scalars.
Options
Optimization Objective and Controls
The default optimization objective is to maximize the first argument of the subroutine. You may use the following optimization options to change the optimization objective and to specify the coefficients (control parameters):
max [=integer] | Maximize the subroutine objective or sum of the values of the subroutine objective (default). By default the first argument of the subroutine is used as the maximization objective. You may change the objective to another argument by specifying an integer argument location. |
min [=integer] | Minimize the subroutine objective or sum of the values of the subroutine objective. By default the first argument of the subroutine is used as the minimization objective. You may change the objective to another argument by specifying an integer argument location. |
ls [=integer] | Perform least squares minimization of the sum of squared values of the subroutine objective. (The objective argument cannot be a scalar value when using this option.) By default the first argument of the subroutine is used as the minimization objective. You can change the objective to another argument by specifying an integer argument location. |
ml [=integer] | Perform maximum likelihood estimation of the sum of the values of the subroutine objective. (The objective argument cannot be a scalar value when using this option.) By default the first argument of the subroutine is used as the maximization objective. You can change the objective to another argument by specifying an integer argument location. Note that the “ml” option specifies the same optimization as when using the “max” option, but permits a different set of Hessian matrix choices. |
coef=integer (default = 2) | Specify the argument number of the function that contains the coefficient controls for the optimization. If the argument is a vector or matrix, each element of the vector or matrix will be treated as a coefficient. If the argument is a series, each element of the series within the current workfile sample will be treated as a coefficient. The default value is 2 so that the second argument is assumed to contain the coefficient controls. |
Optimization Options
grads=integer | Specifies an argument number corresponding to analytic gradients for each of the coefficients. If this option is not specified, gradients are evaluated numerically. Available for “ls” and “ml” estimation. • If the objective argument is a scalar, the gradient argument should be a vector of length equal to the number of elements in the coefficient argument. • If the objective argument is a series, the gradient argument should be a group object containing one series per element of the coefficient argument. The series observations should contain the corresponding derivatives for each observation in the current workfile sample. • For a vector objective, the gradient argument should be a matrix with number of rows equal to the length of the objective vector, and columns equal to the number of elements in the coefficient argument. • “grad=” may not be specified if the objective is a matrix. |
hess=arg | Specify the type of Hessian approximation: “numeric” (numerical approximation), “bfgs” (Broyden–Fletcher–Goldfarb–Shanno), or “opg” (outer product of gradients, or BHHH). “opg” is only available when using “ls” or “ml” optimization. The default value is “bfgs” unless using “ls” optimization, which defaults to “opg”. |
step=arg (default_= “marquardt”) | Set the step method: “marquardt”, “dogleg” or “linesearch”. |
scale=arg | Set the scaling method: “maxcurve” (default), or “none”. |
m=int | Set the maximum number of iterations |
c=number | Set the convergence criteria. |
trust=number (default=0.25) | Sets the initial trust region size as a proportion of the initial control values. Smaller values of this parameter may be used to provide a more cautious start to the optimization in cases where larger steps immediately lead into an undesirable region of the objective. Larger values may be used to reduce the iteration count in cases where the objective is well behaved but the initial values may be far from the optimum values. |
deriv=high | Always use high precision numerical derivatives. Without this option, EViews will start by using lower precision derivatives, and switch to higher precision as the optimization progresses. |
feps=number (default=2.2e-16) | Set the expected relative accuracy of the objective function. The value indicates what fraction of the observed objective value should be considered to be random noise. |
noerr | Turn off error reporting. |
finalh=name | Save the final Hessian matrix into the workfile with name name. For “hess=bfgs”, the final Hessian will be based on numeric derivatives rather than the BFGS approximation used during the optimization since the BFGS approximation need not converge to the true Hessian. |
Examples
The first example estimates a regression model using maximum likelihood. The subroutine LOGLIKE first computes the regression residuals using the coefficients in the vector BETA along with the dependent variable series given by DEP and the regressors in the group REGS.
subroutine loglike(series logl, vector beta, series dep, group regs)
series r = dep - beta(1) - beta(2)*regs(1) - beta(3)*regs(2) - beta(4)*regs(3)
logl = @log((1/beta(5))*@dnorm(r/beta(5)))
endsub
series LL
vector(5) MLCoefs
MLCoefs = 1
MLCoefs(5) = 100
optimize(ml=1, finalh=mlhess, hess=numeric) loglike(LL, MLCoefs, y, xs)
The optimize command instructs EViews to use the LOGLIKE subroutine for purposes of maximization, and to use maximum likelihood to maximize the sum (over the workfile sample) of the LL series with respect to the five elements of the vector MLCOEFS. EViews employs a numeric approximation to the Hessian in optimization, and saves the final estimate in the workfile in the sym object MLHESS.
Notice that we specify initial values for the MLCOEFS coefficients prior to calling the optimization routine.
Our second example recasts the estimation above as a least squares optimization problem, and illustrates the use of the “grads=” option to employ analytically computed gradients defined in the subroutine.
subroutine leastsquareswithgrads(series r, vector beta, group grads, series dep, group regs)
r = dep - beta(1) - beta(2)*regs(1) - beta(3)*regs(2) - beta(4)*regs(3)
grads(1) = 1
grads(2) = regs(1)
grads(3) = regs(2)
grads(4) = regs(3)
endsub
series LSresid
vector(4) LSCoefs
lscoefs = 1
series grads1
series grads2
series grads3
series grads4
group grads grads1 grads2 grads3 grads4
optimize(ls=1, grads=3) leastsquareswithgrads(LSresid, lscoefs, grads, y, xs)
Note that for a linear least squares problem, the derivatives of the coefficients are trivial - the regressors themselves (and a series of ones for the constant).
The next example uses matrix computation to obtain an optimizer objective that finds the solution to the same least squares problem. While the optimizer is not a solver, we can trick it into solving that equation by creating a vector of residuals equal to
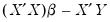
, and asking the optimizer to find the values of
that minimize the square of those residuals:
subroutine local matrixsolve(vector rvec, vector beta, series dep, group regs)
stom(regs, xmat)
xmat = @hcat(@ones(100), xmat)
stom(dep, yvec)
rvec = @transpose(xmat)*xmat*beta - @transpose(xmat)*yvec
rvec = @epow(rvec,2)
endsub
vector(4) MSCoefs
MSCoefs = 1
vector(4) rvec
optimize(min=1) matrixsolve(rvec, mscoefs, y, xs)
The first few lines of the subroutine convert the input dependent variable series and regressor group into matrices. Note that the regressor group does not contain a constant term upon input, so we append a column of ones to the regression matrix XMAT, using the @hcat command.
Lastly, we define a subroutine containing the quadratic form, and use the optimize command to find the value that minimizes the function:
subroutine f(scalar !y, scalar !x)
!y = 5*!x^2 - 3*!x - 2
endsub
scalar in = 0
scalar out = 0
optimize(min) f(out, in)
The subroutine F calculates the simple quadratic formula:
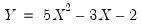 | (17.1) |
which attains a minimum of -2.45 at an IN value of 0.3.
Cross-references
Save the current EViews global options settings “.INI” files into a directory.
Syntax
optsave directory
Save a copy of the current global options settings into the specified directory. Usually this command will be used in conjuncture with a later use of the optset command. Any existing option settings in the directory will be overwritten.
“General Options” and “Graphics Defaults” will always be saved. “Database registry settings” and “Database object aliases” will only be saved if the file location setting for the “Database Registry” and “Alias Map Path” is the same as the file location of the INI File Path.
If the directory name is omitted, the option settings currently in effect will be used to replace the default global options. (This can be used to copy option settings back into your default settings after the optset command has switched to using options in a different directory).
Note that this command does not change which set of options are active. You must follow this command with the optset command if you would like to switch to using the saved copy as your active set of options.
Cross-references
See
Appendix A. “Global Options” for discussion of the global options.
Replace the current EViews global options settings “.INI” files with ones based in a different directory.
Syntax
optset directory
Temporarily switch to using the global options settings stored within “.INI” files in the specified directory. These will typically have been previously saved by using the optsave command.
“General Options” and “Graphics Defaults” will always be switched. “Database registry settings” and Database object aliases” will only be switched if the file location setting for the “Database Registry” and “Alias Map Path” is the same as the file location of the INI File Path.
The new options will stay in effect until EViews is restarted or until the optset command is executed again with a different directory. After the optset command has been issued, changing settings using the menu will modify settings in the new directory.
If the directory name is omitted, the global options settings will be reset to use the settings from the default location (the same as restarting EViews).
Note that you can use the command “optset .\” in a program to switch to using the global options saved in the same directory as the program file. This can be used to ensure that multiple users always use the same global options settings when running a shared program.
Cross-references
See
Appendix A. “Global Options” for discussion of the global options.
Estimation of ordered dependent variable models.
Syntax
equation name.ordered(options) y x1 [x2 x3 ...]
equation name.ordered(options) specification
Options
d=arg (default=“n”) | Specify likelihood: normal likelihood function, ordered probit (“n”), logistic likelihood function, ordered logit (“l”), Type I extreme value likelihood function, ordered Gompit (“x”). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method)., “glm” (GLM method).. |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of preliminary EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
If you choose to employ user specified starting values, the parameters corresponding to the limit points must be in ascending order.
Examples
ordered(d=l,cov=huber) y c wage edu kids
estimates an ordered logit model of Y on a constant, WAGE, EDU, and KIDS with QML standard errors. This command uses the default quadratic hill climbing algorithm.
param c(1) .1 c(2) .2 c(3) .3 c(4) .4 c(5).5
equation eq1.binary(s) y c x z
coef betahat = @coefs
estimates an ordered probit model of Y on a constant, X, and Z from the specified starting values. The estimated coefficients are then stored in the coefficient vector BETAHAT.
Cross-references
See
Equation::ordered for the equivalent equation object command.
Redirect printer output.
You may specify that any procedure that would normally send output to the printer puts output in a text, Rich Text Format (RTF), or comma-separated value (CSV) file, in a spool object, or into frozen table or graph objects in the current workfile.
Syntax
output base_name
output(f) base_name
output(options) [path\]file_name
output(s) spool_name
output off
By default, the output command redirects the output into frozen objects. You should supply a base name after the output keyword. Each subsequent print command will create a new table or graph object in the current workfile, using the base name and an identifying number. For example, if you supply the base name of “OUT”, the first print command will generate a table or graph named OUT01, the second print command will generate OUT02, and so on.
You can also use the optional settings, described below, to redirect table and text output to a text, RTF, or CSV file, or all output (including graphs) to an RTF file. If you elect to redirect output to a file, you must specify a filename.
You can use the “s” option, described below, to redirect all output to a spool object.
When followed by the optional keyword off, the output command turns off output redirection. Subsequent print commands will be directed to the printer.
Options
f | Redirect all output to frozen objects in the default workfile, using base_name. |
t | Redirect table and text output to a text file. Graphic output will still be sent to the printer. |
r | Redirect all output to an Rich Text Format (RTF) file. |
v | Redirect all output to an comma-separated value (CSV) file. |
s | Redirect all output to a spool object. |
o | Overwrite file if necessary. If the specified filename for the text, RTF, or CSV file exists, overwrite the file. The default is to append to the file. Only Applicable when text, RTF, or CSV file output (specified using options “t”, “r”, or “v”). |
c | Command logging. Output both the output, and the command used to generate the output. This option is only applicable in a program when used in conjunction with the
pon command which enables automatic printing of output. The use of pon is not required when output is specified in a command window. Only Applicable when text, RTF, or CSV file output (specified using options “t”, “r”, or “v”). |
Examples
output print_
causes the first print command to generate a table or graph object named PRINT_01, the second print command to generate an object named PRINT_02, and so on.
output(t) c:\data\results
equation eq1.ls(p) log(gdp) c log(k) log(l)
eq1.resids(g,p)
output off
The first line redirects printing to the RESULTS.TXT file, while the print option of the second and third lines sends the graph output to the printer. The last line turns output redirection off and restores normal printer use.
If instead, the first line read:
output(r) c:\data\results
all subsequent output would be sent to the RTF file RESULTS.RTF.
output(s) mySpool
redirects all output to the MYSPOOL spool. If the spool already exists, printed objects will be appended to the end of the spool.
Cross-references
See
“Print Setup” for further discussion.
Append contents of the specified workfile page to the active workfile page.
Syntax
pageappend(options) wfname[\pgname] [object_list]
where wfname is the name of a workfile that is currently in memory. You may optionally provide the name of a page in wfname that you wish to used as a source, and the list of objects to be read from the source workfile page. If no wfname is provided, EViews will use the default page in the source workfile.
The command appends the contents of the source page to the active page in the default workfile. The target page is first unstructured (if necessary) and its range is expanded to encompass the combined range of the sample from the source workfile page, and the destination page.
The default behavior is to append all series and alpha objects (but not other types) from the source page, but the optional object_list may be provided to specify specific series, or to specify objects other than series or alpha objects to be included. Command options may also be used to modify the list of objects to be included.
Note that since this operation is performed in place, the original workfile page cannot be recovered. We recommend that you consider backing up your original page using
pagecopy.
Options
smpl=smpl_spec | Specifies an optional sample identifying which observations from the source page are to be appended. Either provide the sample range in double quotes or specify a named sample object. The default is “@all”. |
allobj | Specifies that all objects (including non-series and non-alpha objects) should be appended. For objects other than series and alphas, appending involves simply copying the objects from the source page to the destination page. This option may not be used with an explicit object_list specification. |
match | Specifies that only series and alphas in the append page that match series and alphas of the same name in the active page should be appended. This option may not be used with “allobj” or with an explicit object_list specification. |
sufix=suffix_arg (default=“_a”) | Specifies a string to be added to the end of the source object name, if necessary, to avoid name collision when creating a new object in the target page. |
obsid=arg | Provides the name of a series used to hold the date or observation ID of each observation in the destination workfile. |
wfid=arg | Provides the name of a (boolean) series to hold an indicator of the source for each observation in the destination workfile (0, if from the destination; 1, if from the source). |
Examples
pageappend updates
appends, to the default workfile page, all of observations in all of the series in the active page of the workfile UPDATES.
pageappend(match, smpl="1999 2003") updates
restricts the series to those which match (by name) those in the default workfile page, and restricts the observations to merge to those between 1999 and 2003.
pageappend newdat\page1 income cons
takes only the INCOME and CONS series from the PAGE1 of the NEWDATA workfile, and appends them to the current workfile page.
pageappend(alltypes, suffix="_1") mydata
appends all objects from MYDATA, merging series with matching names, and renaming other matching objects by appending “_1” to the end of the name.
Cross-references
Contract the active workfile page according to the specified sample.
Syntax
pagecontract smpl_spec
where
smpl_spec is a sample specification. Contraction removes observations not in specified sample from the active workfile page. Note that since this operation is performed in place, you may wish to backup your original page (see
pagecopy) prior to contracting.
Examples
pagecontract if income<50000 and race=2
removes all observations with INCOME values greater than or equal to 50000 and RACE not equal to 2.
pagecontract 1920 1940 1945 2000
removes observations for the years 1941 to 1944.
Cross-references
Copies all or part of the active workfile page to a new workfile, or to a new page within the default workfile.
Syntax
pagecopy(options) [object_list]
where the optional object_list specifies the workfile objects to be copied. If object_list is not provided, all objects will be copied subject to the option restrictions discussed below.
If copying objects to a new page within the default workfile, you may choose to copy series objects (series, alphas, and links) by value or by link (by employing the “bylink” option). If you elect to copy by value, links in the source page will converted to ordinary series and alpha objects when they are copied. If you copy by link, series and alpha objects in the source page are converted to links when copied. The default is to copy by value.
If you copy objects to a new workfile, data objects must be copied by value.
Options
bylink | Specifies that series and alpha objects be copied as links to the source page. This option is not available if you use the “wf=” option, since linking requires that the destination page be in the same workfile as the source page. Automatically sets the “dataonly” option so that only series, alphas, links, and valmaps will be copied. |
smpl=smpl_spec | Specifies an optional sample identifying which observations from the source page are to be appended. Either provide the sample range in double quotes or specify a named sample object. The default is “@all”. |
rndobs=integer | Copy only a random subsample of integer observations from the specified sample. Not available with “bylink,” “rndpct,” or “prob.” |
rndpct=arg | Copy only a random percentage subsample of arg (a number between 0 and 100) of the specified sample. Not available with “bylink,” “rndobs,” or “prob.” |
prob=arg | Copies a random subsample where each observation has a fixed probability, prob, of being selected. prob should be entered as a percentage value (a number between 0 and 100). Not available with “bylink,” “rndobs,” or “rndpc” |
dataonly | Only series, alphas, links, and valmaps should be copied. The default is to copy all objects (unless the “bylink” option is specified, in which case only series objects are copied). |
nolinks | Do not copy links from the source page. |
wf=wf_name | Optional name for the destination workfile. If not provided, EViews will create a new untitled workfile. Not available if copying using the “bylink” option. |
page=page_name | Optional name for the newly created page. If not provided, EViews will use the next available name of the form “Untitled##”, where ## is a number. |
Examples
pagecopy(page=allvalue, wf=newwf)
will first create a new workfile named NEWWF, with a page ALLVALUE that has the same structure as the current page. Next, all of the objects in the active workfile page will be copied into the new page, with the series objects copied by value. In contrast,
pagecopy(bylink, page=alllink)
will instead create a page ALLLINK in the existing workfile, and will copy all series objects by creating links in the new page.
pagecopy(page=partcopy, bylink, smpl="1950 2000 if gender=""male""") a* *z
will create a new page named PARTCOPY in the existing workfile containing the specified subsample of observations, and will copy all series objects in the current page beginning with the letter “A” or ending with the letter “Z”. The objects will be copied by creating links in the new page.
pagecopy(page=rndcopy, smpl="1950 2000 if gender=""male""", rndobs=200, dataonly, nolinks)
creates a new workfile and page RNDCOPY containing a 200 observation random sample from the specified subsample. Series and alpha objects only will be copied by value from the source page.
Cross-references
Create a new page in the default workfile. The new page becomes the active page.
Syntax
pagecreate(options) freq[(subperiod_opts)] start_date end_date [num_cross_sections]
pagecreate(options) u num_observations
pagecreate(id_method[,options]) id_list [@srcpage page_list]
pagecreate(idcross[,options]) id1 id2 [@srcpage page1 page2]
pagecreate(idcross[,options]) id1 @range(freq, start_date, end_date) [@srcpage page1]
These different forms of the pagecreate command encompass three distinct approaches to creating a new workfile page: (1) regular frequency description or unstructured data description; (2) using the union or intersection of unique values from one or more ID series in existing workfile pages; (3) using the cross of unique values from two identifier series or from an identifier series and a date range. Each of these approaches is described in greater detail below.
Regular Frequency or Unstructured Description
The first two forms of the command permit you to create a new workfile page using a regular frequency or unstructured description:
• pagecreate(options) freq[(subperiod_opts)] start_date end_date [num_cross_sections]
• pagecreate(options) u num_observations
The first form of the command should be employed to create a regular frequency page with the specified frequency, start, and end date. If you include the optional argument
num_cross_sections, EViews will create a balanced panel page using integer identifiers for each of the cross-sections. Note that more complex panel structures may be defined using
pagestruct.
The second form of the command creates an unstructured workfile with the specified number of observations.
Note that these forms of the command are analogous to
wfcreate except that instead of creating a new workfile, we create a new page in the default workfile.
The freq argument should be specified using one of the following forms:
Sec[opt], 5Sec[opt], 15Sec[opt], 30Sec[opt] | Seconds in intervals of: 1, 5, 15, or 30 seconds, respectively. You may optionally specify days of the week and start and end times during the day using the opt parameter. See explanation of subperiod options below. |
Min[opt], 2Min[opt], 5Min[opt], 10Min[opt], 15Min[opt], 20Min[opt], 30Min[[opt] | Minutes in intervals of: 1, 2, 5, 10, 15, 20, or 30 minutes, respectively. You may optionally specify days of the week and start and end times during the day using the opt parameter. See explanation of subperiod options below. |
H[opt], 2H[opt], 4H[opt], 6H[opt], 8H[opt], 12H[[opt] | Hours in intervals of: 1, 2, 4, 6, 8, or 12 hours, respectively. You may optionally specify days of the week and start and end times during the day using the opt parameter. See explanation of subperiod options below. |
D(s, e) | Daily with arbitrary days of the week. Specify the first and last days of the week with integers s and e, where Monday is given the number 1 and Sunday is given the number 7. (Note that the “D” option used to specify a 5-day frequency in versions prior to EViews 7). |
D5 or 5 | Daily with five days per week, Monday through Friday. |
D7 or 7 | Daily with seven days per week. |
W | Weekly |
T | Ten-day (daily in intervals of ten). |
F | Fortnight |
BM | Bimonthly |
M | Monthly |
Q | Quarterly |
S | Semi-annual |
A or Y | Annual |
2Y, 3Y, 4Y, 5Y, 6Y, 7Y, 8Y, 9Y, 10Y, 20Y | Multi-year in intervals of: 2, 3, 4, 5, 6, 7, 8, 9, 10, or 20 years, respectively. |
Subperiod options
EViews allows for setting the days of the week and the time of day within intraday frequencies, which include s, , and . For instance, you may specify hourly data between 8AM and 5PM on Monday through Wednesday. These subperiod options should follow the frequency keyword and be enclosed in parentheses.
To specify days of the week, use integers to indicate the days, where Monday is given the number 1 and Sunday is given the number 7. For example,
pagecreate(wf=strhours) 30MIN(1-6, 8:00-17:00) 1/3/2000 12/30/2000
indicates a half-hour frequency that includes Monday through Saturday from 8AM to 5PM.
To specify the start and end times, you may use either a 24 hour clock, including minutes and optionally seconds, or a 12 hour clock using AM and PM. For example, each of the following represents 8PM: 8PM, 8:00PM, 8:00:00PM, 20:00, and 20:00:00. Thus, our previous example could have been written:
pagecreate(wf=strhours) 30MIN(1-6, 8AM-5PM) 1/3/2000 12/30/2000
Note that day and time ranges may be delimited by either commas or dashes. So this command is also equivalent to:
pagecreate(wf=strhours) 30MIN(1,6, 8AM,5PM) 1/3/2000 12/30/2000
though you will likely find the dashes easier to read.
If you wish to include all days of the week but would like to specify a start and end time, set the date range to include all days and then specify the times. The day of the week parameter appears first and is required if you wish to supply the time of day parameters. For instance,
pagecreate(wf=storehours) 30MIN(1-7, 10AM-3PM) 1/3/2000 12/30/2000
indicates a half-hour frequency from 10AM to 3PM on all days of the week.
You may also include a time with the start and end date parameters to specify partial days at the beginning or end of the workfile. For example,
pagecreate(wf=strhours) 30MIN(1-6, 8AM-5PM) 1/3/2000 10AM 12/30/2000 2PM
creates the same workfile page as above, but limits the first day, 1/3/2000, to 10AM - 5PM and the last day, 12/30/2000, to 8AM - 2PM.
Unique Values from a Set of Identifier Series
The next form of the command allows for creating pages from the unique values of one or more identifier series found in one or more workfile pages:
• pagecreate(id_method[,options]) identifier_list [@srcpage page_list]
The identifier_list should include one or more ID series. If more than one ID series is provided, EViews will use the values that are unique across all of the series. If you wish to create a page with a date structure, you should specify one of your identifiers using the special “@DATE” keyword identifier, enclosing the series (or the date ID component series) inside parentheses. If you wish to use the date ID values from the source workfile page, you may use the “@DATE” keyword without arguments.
The id_method describes how to handle unique ID values that differ across multiple pages:
id | Use the observed values of the series in the identifier_list in specified page. |
idunion | Use the union of the observed values of the series in the identifier_list in the specified pages. |
idintersect | Use the intersection of the observed values of the series in the identifier_list in the specified pages. |
If the optional source page or list of source pages is not provided, EViews will use the default workfile page. Note that if a single workfile page is used, the two ID methods yield identical results.
Cross of Unique Values from Two Identifier Series or from an Identifier Series and a Date Range
The last two forms of the command create a new page by crossing the unique values in two ID series located in one or more pages, or by crossing an ID series from one page with a date range. First, you may specify a pair of identifiers, and optionally source pages where they are located,
• pagecreate(idcross[,options]) id1 id2 [@srcpage page1 page2]
You may instruct EViews to create a date structured page by specifying one of your two identifiers using a “@DATE” specification as described above.
Alternately, you may provide a single identifier and a range specification using the “@RANGE” keyword with a freq, start_date, and end_date, and optionally, the location of the identifier series.
• pagecreate(idcross[,options]) id1 @range(freq, start_date, end_date) [@srcpage page1]
Options
smpl=smpl_spec | Specifies an optional sample identifying which observations to use when creating a page using the id_method option. Either provide the sample range in double quotes or specify a named sample object. The default is “@all”. When multiple source workfiles are involved, the specified sample must be valid for all workfiles. |
page=page_name | Optional name for the newly created page. If not provided, EViews will use the next available name of the form “Untitled##”, where ## is a number. |
wf=wf_name | Optional name for the new workfile. If not provided, EViews will create a new page in the default workfile. |
prompt | Force the dialog to appear from within a program. |
Examples
Regular Frequency or Unstructured Description
The two commands:
pagecreate(page=annual) a 1950 2005
pagecreate(page=unstruct) u 1000
create new pages in the existing workfile. The first page is an annual page named ANNUAL, containing data from 1950 to 2005; the second is a 1000 observation unstructured page named UNSTRUCT.
pagecreate(page=mypanel) a 1935 1954 10
creates a new workfile page named MYPANEL, containing a 10 cross-section annual panel for the years 1935 to 1954.
pagecreate(page=fourday) D(1,4) 1/3/2000 12/31/2000
specifies a daily workfile page from January 3, 2000 to December 31, 2000, including only Monday through Thursday. The day range may be delimited by either a comma or a dash, such that
pagecreate(wf=fourday) D(1-4) 1/3/2000 12/31/2000
is equivalent to the previous command.
pagecreate(wf=captimes) 15SEC(2-4) 1/3/2000 12/30/2000
creates a workfile page with 15 second intervals on Tuesday through Thursday only, from 1/3/2000 to 12/30/2000.
Unique Values from a Set of Identifier Series
pagecreate(id, page=statepage) state
creates a new page STATEIND using the distinct values of STATE in the current workfile page.
pagecreate(id, page=statepage) state industry
creates a new page named STATEIND, using the distinct STATE/INDUSTRY values in the active page.
pagecreate(id, page=stateyear) state @date(year)
pagecreate(id, page=statemonth) @date(year, month)
use STATE, along with YEAR, and the YEAR and MONTH series respectively, to form identifiers that will be used in creating the new dated workfile pages.
pagecreate(id, smpl="if sex=1") crossid @date
creates a new page using CROSSID and existing date ID values of the active workfile page. Note that only observations in the subsample defined by “@all if sex=1” are used to determine the unique values.
pagecreate(id, page=AllStates, smpl="if sex=""Female""") stateid @srcpage north south east west
creates a new page ALLSTATES structured using the union of the unique values of STATEID from the NORTH, SOUTH, EAST and WEST workfile pages that are in the sample “if sex="Female"”. Note the use of the double quote escape character for the double quotes in the sample string.
pagecreate(idintersect, page=CommonStates, smpl="1950 2005") stateid @srcpage page1 page2 page3
creates a new page name COMMONSTATES structured using the intersection of the unique values of STATEID taken from the pages PAGE1, PAGE2, and PAGE3.
Cross of Unique Values from Two Identifier Series or from an Identifier Series and a Date Range
pagecreate(idcross,page=UndatedPanel) id1 id2 @srcpage page1 page2
will add the new page UNDATEDPANEL to the current workfile. UNDATEDPANEL will be structured as an undated panel using values from the cross of ID1 from PAGE1 and ID2 from PAGE2.
To create a dated page using the “idcross” option, you must tag one of the identifiers using an “@DATE” specification:
pagecreate(idcross,page=AnnualPanel) id1 @date(year) @srcpage page1 page2
You may also specify the cross of an identifier with a date range:
pagecreate(idcross,page=QuarterlyPanel) id1 @range(q, 1950, 2006) @srcpage page1
creates a quarterly panel page named QUARTERLYPANEL using the cross of ID1 taken from PAGE1, and quarterly observations from 1950q1 to 2006q4.
Cross-references
Delete the named pages from the default workfile.
Syntax
pagedelete [pgname1 pgname2 pgname3...]
By default pagedelete will delete the currently active page in the default workfile. You may specify other pages to delete by listing them, in a space delimited list, after the pagedelete command.
Examples
pagedelete page1 page2
Cross-references
Load one or more new pages in the default workfile.
Syntax
pageload [path\]workfile_name [page1] [page2] [...]
pageload(options) source_description [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@selectif condition]
pageload(options)source_description table_description [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@selectif condition]
The basic syntax for
pageload follows that of
wfopen. The difference between the two commands is that pageload creates a new page in the default workfile, rather than opening or creating a new workfile. If a page is loaded with a name that matches an existing page, EViews will rename the new page to the next available name (
e.g., “INDIVID” will be renamed “INDIVID1”.
If a workfile is provided as the source file, EViews will, by default, open all pages in the source workfile. Specific pages may be loaded by providing their names.
Examples
pageload "c:\my documents\data\panel1"
loads the workfile PANEL1.WF1 from the specified directory. All of the pages in the workfile will be loaded as new pages into the current workfile.
pageload f.wf1 mypage
loads the page “mypage” in the workfile F.WF1 located in the default directory.
See the extensive set of examples in
wfopen.
Cross-references
Refresh all links and auto-series in the active workfile page. Primarily used to refresh links that use external database data.
Syntax
pagerefresh
Cross-references
See also
“Series Links” for a description of link objects, and
“Auto-Updating Series” for a discussion of auto-updating series.
See
wfrefresh to reference an entire workfile.
Rename the specified workfile page.
Syntax
pagerename old_name new_name
renames the old_name page in the default workfile to new_name. Page names are case-insensitive for purposes of comparison, even though they are displayed using the input case.
Examples
pagerename Page1 StateByYear
Cross-references
Save the active page in the default workfile as an EViews workfile (.WF1 file) or as a foreign data source.
Syntax
pagesave(options) [path\]filename
pagesave(options) source_description [nonames] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@smpl smpl_spec]
pagesave(options) source_description table_description [nonames] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@smpl smpl_spec]
The command saves the active page in the specified directory using filename. By default, the page is saved as an EViews workfile, but options may be used to save all or part of the page in a foreign file or data source.
When saving to a foreign data file, the basic specification consists of a “type=” option and source_description and table_description arguments which specify the format of the foreign data file. See below for details on source_description and table_description.
The remaining optional elements specify the actual elements to be saved.
Options
type=arg, t=arg | Optional type specification: (see table below). ODBC support is not provided in EViews Standard Edition. |
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If a “mode=” option is not provided, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
maptype=arg | Write selected maps as: numeric (“n”), character (“c”), both numeric and character (“b”). |
nomapval | Do not write mapped values for series with attached value labels (the default is to write mapped values) |
noid | Do not write observation identifiers to foreign data files (by default, EViews will include a column with the date or observation identifier). |
nonames | Do not write variable names (only application to file formats that support writing raw data without variable names). Only available in EViews 12 and later. EViews 11 and older should use nonames as an argument after the output file name. |
attr | Include object attributes (if the output type supports it). When specified, the first column will contain attribute names and each attribute value will be displayed after the name row. |
Excel Options
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If the “mode=” option is not used, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
Excel 2007 Options
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If the “mode=” option is not used, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
cellfmt=arg | Specify whether to use EViews, pre-existing, or remove cell formatting (colors, font, number formatting when possible, column widths and row heights) for the written range. arg may be “eviews” (replace current formatting in the file with the same cell formatting in EViews), “preserve” (leave current cell formatting already in the Excel file), or “clear” (remove current formatting and do not replace). |
strlen=arg (default = 256) | Specify the maximum the number of characters written for cells containing text. Strings in cells which are longer the max, will be truncated. |
The following table summaries the various foreign formats, along with the corresponding “type=” keywords:
| | |
Access | “access” | |
Aremos-TSD | “a”, “aremos”, “tsd” | |
Binary | “binary” | |
dBASE | “dbase” | |
Excel (through 2003) | “excel” | Yes |
Excel 2007 (xml) | “excelxml” | Yes |
EViews Workfile | --- | |
Gauss Dataset | “gauss” | |
GiveWin/PcGive | “g”, “give” | |
HTML | “html” | |
JSON** | json | |
Lotus 1-2-3 | “lotus” | |
ODBC Dsn File | “dsn” | |
ODBC Data Source | “odbc” | |
MicroTSP Workfile | “dos”, “microtsp” | |
MicroTSP Mac Workfile | “mac” | |
RATS 4.x | “r”, “rats” | |
RATS Portable / TROLL | “l”, “trl” | |
SAS Program | “sasprog” | |
SAS Transport | “sasxport” | |
SPSS | “spss” | |
SPSS Portable | “spssport” | |
Stata (Version 7 Format) | “stata” | |
Tableau Data Extract | “tde” | |
Text / ASCII | “text” | Yes |
TSP Portable | “t”, “tsp” | |
Note that if you wish to save your Excel 2007 XML file with macros enabled, you should specify the explicit filename extension “.XLSM”.
Foreign Data Descriptions
When saving to a foreign data format the base specification consists of a basic specification of source_description and table_description which specify the exact details of the format.
The command for saving as foreign data formats is
pagesave(options) source_description [table_description] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@smpl smpl_spec] [nonames]
where the syntax of the table_description and variables_description differs slightly depending on the type of file.
• @keep, @drop, @keepmap, @dropmap, and @smpl arguments may be used to control what objects and observations to write.
• The nonames keyword may be used to suppress the writing of variable names in file formats that support writing raw data without variable names.
• Note that the JSON type will ignore any @keep, @drop, and @smpl arguments.
Excel Files
The base syntax for writing Excel files is:
pagesave(options) source_description [table_description]
where source_description is the path and name of the Excel file to be saved, and where the following table_description elements may be employed:
• “range = arg”, where arg is a range of cells to read from the Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the excel workbook to refer to a range or cell may be used to specify the cells to read.
• “byrow”, transpose the incoming data. This option allows you to read files where the series are contained in rows (one row per series) rather than columns.
Examples
HTML Files
The base syntax for saving HTML files is:
pagesave(options) source_description [table_description]
where source_description is the path and name of the file to be saved, and where the following table_description element may be employed:
• “byrow”, transpose the data. This option allows you to write files where the series are contained in rows (one row per series) rather than columns.
Text and Binary and Other Files
The base syntax for saving other files is:
pagesave(options) source_description
where source_description is the path and name of the file to be saved.
Examples
EViews Workfile Examples
pagesave new_wf
saves the current EViews workfile page as “New_wf.WF1” in the default directory.
pagesave "c:\documents and settings\my data\consump"
saves the current workfile page as “Consump.WF1” in the specified path.
pagesave macro @keep gdp unemp
saves the two series GDP and UNEMP in a separate workfile, “macro.WF1” in the default directory.
pagesave macro @dropmap gdp*
saves all of the series in the current workfile, other than those that match the name pattern “gdp*” in a workfile, “macro.WF1” in the default directory.
The command:
pagesave "<mydropboxdrive>"\folder\nipa.wf1"
will save the file to the cloud location MYDROPBOXDRIVE.
Foreign Data Examples
pagesave(type=excelxml, mode=update) macro.xlsx
saves the current workfile page as a modern Excel “.XLSX” file.
pagesave(type=excelxml, mode=update) macro.xlsx range="Sheet2!a1" byrow @keep gdp unemp
will save the two series GDP and UNEMP into the existing Excel file “macro.XLSX”, specifying that the series should be written by row, starting in cell A1 on sheet Sheet2.
To save the latter file in a macro-enabled Excel 2007 file, you should specify the explicit filename extension “.XLSM”:
pagesave(type=excelxml, mode=update) macro.xlsm range="Sheet2!a1" byrow @keep gdp unemp
Alternately,
pagesave(type=excelxml, noid) macro.xlsx range="Sheet2!a1"
will save the current workfile page as the Excel file “macro.XLSX” but will not include a column of dates.
If you wish to save a column of dates in a specific date format, you can do so by first creating an alpha series in the workfile with the specified format, then saving the file with the “noid” option including that alpha series:
alpha mydates = @datestr(@date, "YYYY-MM-DD")
pagesave(type=excelxml, noid) macro.xlsm range="Sheet2!a1" @keep mydates gdp unemp
Will save the series GDP and UNEMP into the Excel file “macro.XLSM” along with a date series with the format “YYYY-MM-DD”.
Cross-references
See also
wfopen and
wfsave.
Make the specified page in the default workfile the active page.
Syntax
pageselect pgname
where pgname is the name of a page in the default workfile.
Examples
pageselect page2
changes the active page to PAGE2.
Cross-references
Sort the current workfile page.
The pagesort command sorts all series in the workfile page on the basis of the values of one or more of the series. For purposes of sorting, NAs are considered to be smaller than any other value. By default, EViews will sort the series in ascending order. You may use options to override the sort order.
EViews will first remove any workfile structures and then will sort the workfile using the specified settings.
Syntax
pagesort(options) arg1 [arg2 arg3…]
List the name of the series or groups by which you wish to sort the workfile. If you list two or more series, pagesort uses the values of the second series to resolve ties from the first series, and values of the third series to resolve ties from the second, and so on.
Options
d | sort in descending order. |
Examples
pagesort(d) inc
sorts all series in the workfile in order of the INC series with the highest value of INC first. NAs in INC (if any) will be placed at the bottom.
pagesort gender race wage
sorts all series in the workfile in order of the values of GENDER from low to high, with ties resolved by ordering on the basis of RACE, with further ties resolved by ordering on the basis of WAGE.
Cross-references
Create a panel structured workfile page using series, alphas, or links from the default workfile page (convert repeated series to repeated observations).
Series in the new panel workfile may be created by stacking series, alphas, and links whose names contain a pattern (series with names that differ only by a “stack identifier”), or by repeating a single series, alpha, or link, for each value in a set of stack identifiers.
Syntax
pagestack(options) stack_id_list [@ series_to_stack]
pagestack(options) pool_name [@ series_to_stack]
pagestack(options) series_name_pattern [@ series_to_stack]
The resulting panel workfile will use the identifiers specified in one of the three forms of the command:
• stack_id_list includes a list of the ID values (e.g., “US UK JPN”).
• pool_name is the name of a pool object that contains the ID values.
• series_name_pattern contains an expression from which the ID values may be determined. The pattern should include the “?” character as a stand in for the parts of the series names containing the stack identifiers. For example, if “CONS?” is the series_name_pattern, EViews will find all series with names beginning with “CONS” and will extract the IDs from the trailing parts of the observed names.
The series_to_stack list may contain two types of entries: stacked series (corresponding to sets of series, alphas, and links whose names contain the stack IDs) and simple series (other series, alphas, and links).
To stack a set of series whose names differ only by the stack IDs, you should enter an expression that includes the “?” character in place of the IDs. You may list the names of a single stacked series (e.g., “GDP?” or “?CONS”), or you may use expressions containing the wildcard character “*” (e.g., “*?” and “?C*”) to specify multiple series.
By default, the stacked series will be named in the new workfile using the base portion of the series name (if you specify “?CONS” the stacked series will be named “CONS”), and will contain the values of the individual series stacked one on top of another. If one of the individual series associated with a particular stack ID does not exist, the corresponding stacked values will be assigned the value NA.
Individual (simple) series may also be stacked. You may list the names of individual simple series (e.g., “POP INC”), or you can specify your series using expressions containing the wildcard character “*” (e.g., “*”, “*C”, and “F*”). A simple series will be stacked on top of itself, once for each ID value. If the target workfile page is in the same workfile, EViews will create a link in the new page; otherwise, the stacked series will contain (repeated) copies of the original values.
When evaluating wildcard expressions, stacked series take precedence over simple series. This means that simple series wildcards will be processed using the list of series not already included as a stacked series.
If the series_to_stack list is not specified, the expression “*? *”, is assumed.
Options
?=name_patt, idreplace = name_patt | Specifies the characters to use instead of the identifier, ”?”, in naming the stacked series. By default, the name_patt is blank, indicating, for example, that the stacked series corresponding to the pattern “GDP?” will be named “GDP” in the stacked workfile page. If pattern is set to “STK”, the stacked series will be named GDPSTK. |
interleave | Interleave the observations in the destination stacked workfile (stack by using all of the series values for the first source observation, followed by the values for the second observation, and so on). The default is to stack observations by identifier (stack the series one on top of each other). |
wf=wf_name | Optional name for the new workfile. If not provided, EViews will create a new page in the default workfile. |
page=page_name | Optional name for the newly created page. If not provided, EViews will use the next available name of the form “Untitled##”, where ## is a number. |
Examples
Consider a workfile that contains the seven series: GDPUS, GDPUK, GDPJPN, CONSUS, CONSUK, CONSJPN, CONSFR, and WORLDGDP.
pagestack us uk jpn @ *?
creates a new, panel structured workfile page with the series GDP and CONS, containing the stacked GDP? series (GDPUS, GDPUK, and GDPJPN) and stacked CONS? series (CONSUS, CONSUK, and CONSJPN). Note that CONSFR and WORLDGDP will not be copied or stacked.
We may specify the stacked series list explicitly. For example:
pagestack(page=stackctry) gdp? @ gdp? cons?
first determines the stack IDs from the names of series beginning with “GDP”, the stacks the GDP? and CONS? series. Note that this latter example also names the new workfile page STACKCTRY.
If we have a pool object, we may instruct EViews to use it to specify the IDs:
pagestack(wf=newwf, page=stackctry) countrypool @ gdp? cons?
Here, the panel structured page STACKCTRY will be created in the workfile NEWWF.
Simple series may be specified by adding them to the stack list, either directly, or using wildcard expressions. Both commands:
pagestack us uk jpn @ gdp? cons? worldgdp consfr
pagestack(wf=altwf) us uk jpn @ gdp? cons? *
stack the various GDP? and CONS? series on top of each other, and stack the simple series GDPFR and WORLDGDP on top of themselves.
In the first case, we create a new panel structured page in the same workfile containing the stacked series GDP and CONS and link objects CONSFR and WORLDGDP, which repeat the values of the series. In the second case, the new panel page in the workfile ALTWF will contain the stacked GDP and CONS, and series named CONSFR and WORLDGDP containing repeated copies of the values of the series.
The following two commands are equivalent:
pagestack(wf=newwf) us uk jpn @ *? *
pagestack(wf=newwf) us uk jpn
Here, every series, alpha, and link in the source workfile is stacked and copied to the destination workfile, either by stacking different series containing the stack_id or by stacking simple series on top of themselves.
The “?=” option may be used to prevent name collision.
pagestack(?="stk") us uk jpn @ gdp? gdp
stacks GDPUS, GDPUK and GDPJPN into a series called GDPSTK and repeats the values of the simple series GDP in the destination series GDP.
Cross-references
For additional discussion, see
“Stacking a Workfile”. See also
pageunstack.
Assign a structure to the active workfile page.
Syntax
pagestruct(options) [id_list]
pagestruct(options) *
where id_list is an (optional) list of ID series. The “*” may be used as shorthand for the indices currently in place.
If an id_list is provided, EViews will attempt to auto determine the workfile structure. Auto-determination may be overridden through the use of options.
If you do not provide an id_list, the workfile will be restructured as a regular frequency workfile. In this case, either the “none” or the “freq=” and “start=” options described below must be provided.
Options
none | Remove the existing workfile structure. |
freq= arg | Specifies a regular frequency; if not provided EViews will auto-determine the frequency. The frequency may be specified as “a” (annual), “s” (semi-annual), “q” (quarterly), “m” (monthly), “w” (weekly), “d” (5-day daily), “7” (7-day daily), or “u” (unstructured/undated). |
start=arg | Start date of the regular frequency structure; if not specified, defaults to “@FIRST”. Legal values for arg are described below. |
end=arg | End date of the regular frequency structure; if not specified, defaults to “@LAST”. Legal values for arg are described below. |
regular, reg | When used with a date ID, this option informs EViews to insert observations (if necessary) to remove gaps in the date series so that it follows the regular frequency calendar. The option has no effect unless a date index is specified. |
create | Allow creation of a new series to serve as an additional ID series when duplicate ID values are found in any group. EViews will use this new series as the observation ID. The default is to prompt in interactive mode and to fail in programs. |
balance=arg, bal=arg | Balance option (for panel data) describing how EViews should handle data that are unbalanced across ID (cross-section) groups. The arg should be formed using a combination of starts (“s”), ends (“e”), and middles (“m”), as in “balance=se” or “balance=m”. If balancing starts (arg contains “s”), EViews will (if necessary) add observations to your workfile so that each cross-section begins at the same observation (the earliest date or observation observed). If balancing ends (arg contains “e”), EViews will add any observations required so that each cross-section ends at the same point (the last date or observation observed). If balancing middles (arg contains “m”) EViews will add observations to ensure that each cross-section has consecutive observations from the earliest date or observation for the cross-section to the last date or observation for the cross-section. Note that “balance=m” is implied by the “regular” option. |
dropna | Specifies that observations which contain missing values (NAs or blank strings) in any ID series (including the date or observation ID) be removed from the workfile. If “dropna” is not specified and NAs are found, EViews will prompt in interactive mode and fail in programs. |
dropbad | Specifies that observations for which any of the date index series contain values that do not represent dates be removed from the workfile. If “dropbad” is not provided and bad dates are present, EViews will prompt in interactive mode and fail in programs. |
The values for start and end dates should contain date literals (actual dates or periods), e.g., “1950q1” and “2/10/1951”, “@FIRST”, or “@LAST” (first and last observed dates in the date ID series). Date literals must be used for the “start=” option when restructuring to a regular frequency.
In addition, offsets from these date specifications can be specified with a “+” or “–” followed by an integer: “@FIRST-5”, “@LAST+2”, “1950m12+6”. Offsets are most often used when resizing the workfile to add or remove observations from the endpoints.
Examples
pagestruct state industry
structures the workfile using the IDs in the STATE and INDUSTRY series.
A date ID series (or a series used to form a date ID) should be tagged using the “@DATE” keyword. For example:
pagestruct state @date(year)
pagestruct(regular) @date(year, month)
A “*” may be used to indicate the indices defined in the current workfile structure.
pagestruct(end=@last+5) *
adds 5 observations to the end of the current workfile.
When you omit the id_list, EViews will attempt to restructured the workfile to a regular frequency. In this case you must either provide the “freq=” and “start=” options to identify the regular frequency structure, or you must specify “none” to remove the existing structure:
pagestruct(freq=a, start=1950)
pagestruct(none)
Cross-references
For extensive discussion, see
“Structuring a Workfile”.
Break links in all link objects and auto-updating series (formulae) in the active workfile page.
You should use some caution with this command as you will not be prompted before the links and auto-updating series are converted.
Syntax
pageunlink
Examples
pageunlink
breaks links in all pages of the active workfile page.
Cross-references
See
“Series Links” for a description of link objects, and
“Auto-Updating Series” for a discussion of auto-updating series.
See
unlink and
wfunlink for object and workfile based unlinking, respectively.
Unstack workfile page (convert repeated observations to repeated series).
Create a new workfile page by taking series objects (series, alphas, or links) in the default workfile page and breaking them into multiple series (or alphas), one for each distinct value found in a user supplied list of series objects. Typically used on a page with a panel structure.
Syntax
pageunstack(options) stack_id obs_id [@ series_to_unstack]
where stack_id is a single series containing the unstacking ID values used to identify the individual unstacked series, obs_id is a series containing the observation IDs, and series_to_unstack is an optional list of series objects to be copied to the new workfile.
Options
namepat =name_pattern | Specifies the pattern from which unstacked series names are constructed, where “*” indicates the original series name and “?” indicates the stack ID. By default the name_pattern is “*?”, indicating, for example, that if we have the IDs “US”, “UK”, “JPN”, the unstacked series corresponding to the series GDP should be named “GDPUS”, “GDPUK”, “GDPJPN” in the unstacked workfile page. |
wf=wf_name | Optional name for the new workfile. If not provided, EViews will create a new page in the default workfile. |
page=page_name | Optional name for the newly created page. If not provided, EViews will use the next available name of the form “Untitled##”, where ## is a number. |
Examples
Consider a workfile that contains the series GDP and CONS which contain the values of Gross Domestic Product and consumption for three countries stacked on top of each other. Suppose further there is an alpha object called COUNTRY containing the observations “US”, “UK”, and “JPN”, which identify which from which country each observation of GDP and CONS comes. Finally, suppose there is a date series DATEID which identifies the date for each observation. The command:
pageunstack country dateid @ gdp cons
creates a new workfile page using the workfile frequency and dates found in DATEID. The page will contain the 6 series GDPUS, GDPUK, GDPJPN, CONSUS, CONSUK, and CONSJPN corresponding to the unstacked GDP and CONS.
Typically the source workfile described above would be structured as a dated panel with the cross-section ID series COUNTRY and the date ID series DATEID. Since the panel has built-in date information, we may use the “@DATE” keyword as the DATEID. The command:
pageunstack country @date @ gdp cons
uses the date portion of the current workfile structure to identify the dates for the unstacked page.
The stack_id must be an ordinary, or an alpha series that uniquely identifies the groupings to use in unstacking the series. obs_id may be one or more ordinary series or alpha series, the combination of which uniquely identify each observation in the new workfile.
You may provide an explicit list of series to unstack following an “@” immediately after the obs_id. Wildcards may be used in this list. For example:
pageunstack country dateid @ g* c*
unstacks all series and alphas that have names that begin with “G” or “C’.
If no series_to_unstack list is provided, all series in the source workfile will be unstacked. Thus, the two commands:
pageunstack country dateid @ *
pageunstack country dateid
are equivalent.
By default, series are named in the destination workfile page by appending the stack_id values to the original series name. Letting “*” stand for the original series name and “?” for the stack_id, names are constructed as “*?”. This default may be changed using the “namepat=” option. For example:
pageunstack(namepat="?_*") country dateid @ gdp cons
creates the series US_GDP, UK_GDP, JPN_GDP, etc.
Cross-references
For additional discussion and examples, see
“Unstacking a Workfile”.
Set parameter values.
Allows you to set the current values of coefficient vectors. The command may be used to provide starting values for the parameters in nonlinear least squares, nonlinear system estimation, and (optionally) ARMA estimation.
Syntax
param coef_name1 number1 [coef_name2 number2 coef_name3 number3…]
List, in pairs, the names of the coefficient vector and its element number followed by the corresponding starting values for any of the parameters in your equation.
Examples
param c(1) .2 c(2) .1 c(3) .5
resets the first three values of the coefficient vector C.
coef(3) beta
param beta(2) .1 beta(3) .5
The first line declares a coefficient vector BETA of length 3 that is initialized with zeros. The second line sets the second and third elements of BETA to 0.1 and 0.5, respectively.
Cross-references
See
“Starting Values” for a discussion of setting initial values in nonlinear estimation.
Line graph.
Provided for backward compatibility. See
line.
Preview objects contained in a database or workfile.
Syntax
Database object preview:
preview(options) [path\]db_name [as shorthand_name]::object_name
Workfile object preview:
preview(options) [path\]workfile name::object_name
preview(options) [path\]workfile name::pagename\object_name
Options
all | Display all data in the series. |
recent | Display recent data in the series. |
log | Display the log of the series. |
pch | Display the percent change of the series data. |
pchy | Display the year-on-year percent change of the series data. |
Examples
preview evdbtest::x
previews object X in database EVDBTEST in the default path.
preview .\temp evdbtest.edb::y
previews object X in database EVDBTEST in the temp folder.
preview(pch,recent) dbpreview::x
previews object X in database EVDBTEST and displays the percentage change of recent data.
preview(pchy) dbpreview::x
previews object X in database EVDBTEST and displays the percentage change.
preview x
previews object X in the active workfile.
preview .\temp\testwf::x
previews object X in workfile TESTWF in the temp folder and displays the log of all the data.
preview(log,all) .\temp\testwf::x
previews object X
preview page1\x
previews object X of page1 in the active workfile.
preview .\temp\testwf::page1\x
previews object X of page1 in workfile TESTWF in the temp folder.
preview(pch,recent) .\temp\testwf::x
previews object X in workfile TESTWF in the temp folder and displays the percentage change of recent data.
Sends views of objects to the default printer.
Syntax
print(options) object1 [object2 object3 …]
print(options) object_name.view_command
print should be followed by a list of object names or a view of an object to be printed. The list of names must be of the same object type. If you do not specify the view of an object, print will print the default view for the object.
Options
p | Print in portrait orientation. |
l | Print in landscape orientation. |
The default orientation is set by clicking on .
Examples
print gdp log(gdp) d(gdp) @pch(gdp)
sends a table of GDP, log of GDP, first difference of GDP, and the percentage change of GDP to the printer.
print graph1 graph2 graph3
prints three graphs on a single page.
To merge the three graphs, realign them in one row, and print in landscape orientation, you may use the commands:
graph mygra.merge graph1 graph2 graph3
mygra.align(3,1,1)
print(l) mygra
To estimate the equation EQ1 and send the output view to the printer.
print eq1.ls gdp c gdp(-1)
Cross-references
See
“Print Setup” for a discussion of print options and the dialog.
See
output for print redirection.
Estimation of binary dependent variable models with normal errors.
Equivalent to “binary(d=n)”.
Declare a program.
Syntax
program [path\]prog_name
Enter a name for the program after the program keyword. If you do not provide a name, EViews will open an untitled program window. Programs are text files, not objects.
Examples
program runreg
opens a program window named RUNREG which is ready for program editing.
Cross-references
See
“EViews Programming” for further details, and examples of writing EViews programs.
Estimate a quantile regression specification.
Syntax
qreg(options) y x1 [x2 x3 ...]
qreg(options) linear_specification
Options
quant=number (default = 0.5) | Quantile to be fit (where number is a value between 0 and 1). |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=arg (default=“sandwich”) | Method for computing coefficient covariance matrix: “iid” (ordinary estimates), “sandwich” (Huber sandwich estimates), “boot” (bootstrap estimates). When “cov=iid” or “cov=sandwich”, EViews will use the sparsity nuisance parameter calculation specified in “spmethod=” when estimating the coefficient covariance matrix. |
bwmethod=arg (default = “hs”) | Method for automatically selecting bandwidth value for use in estimation of sparsity and coefficient covariance matrix: “hs” (Hall-Sheather), “bf” (Bofinger), “c” (Chamberlain). |
bw =number | Use user-specified bandwidth value in place of automatic method specified in “bwmethod=”. |
bwsize=number (default = 0.05) | Size parameter for use in computation of bandwidth (used when “bw=hs” and “bw=bf”). |
spmethod=arg (default=“kernel”) | Sparsity estimation method: “resid” (Siddiqui using residuals), “fitted” (Siddiqui using fitted quantiles at mean values of regressors), “kernel” (Kernel density using residuals) Note: “spmethod=resid” is not available when “cov=sandwich”. |
btmethod=arg (default= “pair”) | Bootstrap method: “resid” (residual bootstrap), “pair” (xy-pair bootstrap), “mcmb” (MCMB bootstrap), “mcmba” (MCMB-A bootstrap). |
btreps=integer (default=100) | Number of bootstrap repetitions |
btseed=positive integer | Seed the bootstrap random number generator. If not specified, EViews will seed the bootstrap random number generator with a single integer draw from the default global random number generator. |
btrnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator for the bootstrap: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
btobs=integer | Number of observations for bootstrap subsampling (when “bsmethod=pair”). Should be significantly greater than the number of regressors and less than or equal to the number of observations used in estimation. EViews will automatically restrict values to the range from the number of regressors and the number of estimation observations. If omitted, the bootstrap will use the number of observations used in estimation. |
btout=name | (optional) Matrix to hold results of bootstrap simulations. |
k=arg (default=“e”) | Kernel function for sparsity and coefficient covariance matrix estimation (when “spmethod=kernel”): “e” (Epanechnikov), “r” (Triangular), “u” (Uniform), “n” (Normal–Gaussian), “b” (Biweight–Quartic), “t” (Triweight), “c” (Cosinus). |
m=integer | Maximum number of iterations. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number (default =0) | Determine starting values for equations. Specify a number between 0 and 1 representing the fraction of preliminary least squares coefficient estimates. Note that out of range values are set to the default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
qreg y c x
estimates the default least absolute deviations (median) regression for the dependent variable Y on a constant and X. The estimates use the Huber Sandwich method for computing the covariance matrix, with individual sparsity estimates obtained using kernel methods. The bandwidth uses the Hall and Sheather formula.
qreg(quant=0.6, cov=boot, btmethod=mcmba) y c x
estimates the quantile regression for the 0.6 quantile using MCMB-A bootstrapping to obtain estimates of the coefficient covariance matrix.
Cross-references
See
“Quantile Regression” for a discussion of the quantile regression.
Reset the workfile range for a regular frequency workfile.
No longer supported. See the replacement command
pagestruct.
Import data from a foreign disk file into series.
May be used to import data into an existing workfile from a text, Excel, or Lotus file on disk.
Unless you need to merge data into an
existing workfile page, we recommend that you use the more powerful, easy-to-use tools for reading data (see
“Creating a Workfile by Reading from a Foreign Data Source”).
See
pageload, and
wfopen for command details.
Syntax
read(options) [path\]file_name name1 [name2 name3 ...]
read(options) [path\]file_name n
You must supply the name of the source file. If you do not include the optional path specification, EViews will look for the file in the default directory. Path specifications may point to local or network drives. If the path specification contains a space, you may enclose the entire expression in double quotation marks.
The input specification follows the source file name. There are two ways to specify the input series. First, you may list the names of the series in the order they appear in the file. Second, if the data file contains a header line for the series names, you may specify the number, n, of series in the file instead of a list of names. EViews will name the n series using the names given in the header line. If you specify a number and the data file does not contain a header line, EViews will name the series as SER01, SER02, SER03, and so on.
To import data into alpha series, you must specify the names of your series, and should enter the tag “$” following the series name (e.g., “NAME $ INCOME CONSUMP”).
Options
prompt | Force the dialog to appear from within a program. |
File type options
t=dat, txt | ASCII (plain text) files. |
t=wk1, wk3 | Lotus spreadsheet files. |
t=xls | Excel spreadsheet files. |
If you do not specify the “t” option, EViews uses the file name extension to determine the file type. If you specify the “t” option, the file name extension will not be used to determine the file type.
Options for ASCII text files
t | Read data organized by series. Default is to read by observation with series in columns. |
na=text | Specify text for NAs. Default is “NA”. |
d=t | Treat tab as delimiter (note: you may specify multiple delimiter options). The default is “d=c” only. |
d=c | Treat comma as delimiter. |
d=s | Treat space as delimiter. |
d=a | Treat alpha numeric characters as delimiter. |
custom = symbol | Specify symbol/character to treat as delimiter. |
mult | Treat multiple delimiters as one. |
name | Series names provided in file. |
label=integer | Number of lines between the header line and the data. Must be used with the “name” option. |
rect (default) / norect | [Treat / Do not treat] file layout as rectangular. |
skipcol = integer | Number of columns to skip. Must be used with the “rect” option. |
skiprow = integer | Number of rows to skip. Must be used with the “rect” option. |
comment= symbol | Specify character/symbol to treat as comment sign. Everything to the right of the comment sign is ignored. Must be used with the “rect” option. |
singlequote | Strings are in single quotes, not double quotes. |
dropstrings | Do not treat strings as NA; simply drop them. |
negparen | Treat numbers in parentheses as negative numbers. |
allowcomma | Allow commas in numbers (note that using commas as a delimiter takes precedence over this option). |
currency= symbol | Specify symbol/character for currency data. |
Options for spreadsheet (Lotus, Excel) files
t | Read data organized by series. Default is to read by observation with series in columns. |
letter_number (default=“b2”) | Coordinate of the upper-left cell containing data. |
s=sheet_name | Sheet name for Excel 5–8 Workbooks. |
Examples
read(t=dat,na=.) a:\mydat.raw id lwage hrs
reads data from an ASCII file MYDAT.RAW in the A: drive. The data in the file are listed by observation, the missing value NA is coded as a “.” (dot or period), and there are three series, which are to be named ID, LWAGE, HRS (from left to right).
read(a2,s=sheet3) cps88.xls 10
reads data from an Excel file CPS88 in the default directory. The data are organized by observation, the upper left data cell is A2, and 10 series are read from a sheet named SHEET3 using names provided in the file.
read(a2, s=sheet2) "\\network\dr 1\cps91.xls" 10
reads the Excel file CPS91 from the network drive specified in the path.
Cross-references
See
“Importing Data” for a discussion and examples of importing data from external files.
Rename an object in the active workfile or database.
Syntax
rename old_name new_name [old_name1 new_name1 [old_name2 new_name2 [...]]]
After the rename keyword, list the pairs of old object names followed by the new names. Note that the name specifications may include matching wildcard patterns.
Examples
rename temp_u u2
renames an object named TEMP_U as U2.
rename aa::temp_u aa::u2
renames the object TEMP_U to U2 in database AA.
rename a* b*
renames all objects beginning with the letter “A” to begin with the letter “B”.
rename a1 a2 b1 b2
renames A1 to A2 and B1 to B2.
Cross-references
See
“Object Basics” for a discussion of working with objects in EViews.
Compute Ramsey’s regression specification error test.
Syntax
reset(n, options)
You must provide the number of powers of fitted terms n to include in the test regression.
Options
prompt | Force the dialog to appear from within a program. |
p | Print the test result. |
Examples
ls lwage c edu race gender
reset(2)
carries out the RESET test by including the square and the cube of the fitted values in the test equation.
Cross-references
See
“Ramsey's RESET Test” for a discussion of the RESET test.
Generate multivariate random normal values.
Fill group of series with multivariate normals random draws.
Syntax
rmvnorm(x, S[, prefix]))
rmvnormc(x, S[, prefix]))
rmvnormi(x, S[, prefix]))
mvnormci(x, S[, prefix]))
where x is an object as described below matrix, and sym S describes the covariance matrix as described below.
• If x doesn't exist, a group object is first created and populated with series named “<prefix>1”, “<prefix>2”, and so on. If omitted, the default prefix is "SER".
• If x is a group, the command fills each observation of the contained series with a draw from the distribution.
• If x is a vector, the command fills the vector with a single draw from the distribution.
• If x is a matrix, the command fills each row of the matrix with a draw from the distribution.
There are four distinct commands in this family which correspond to different interpretations for S. These forms are distinguished by the characters in the command name that follow the initial string “rmvnorm”:
“” | Supply  , the covariance matrix of distribution. |
“c” | Supply the Cholesky decomposition of  . This form is more efficient when performing multiple draws from the same distribution (compute the Cholesky once, but sample many times). |
“i “ | Supply  . This form is more efficient than explicitly inverting 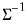 to supply 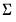 . |
“ic” | Supply the Cholesky decomposition of 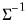 . This form combines the efficiencies of the above forms. |
Examples
sym a = @inner(@mnrnd(100, 10))
matrix(1000, 10) x
rmvnorm(x, a)
rmvnormc(x, @cholesky(a))
rmvnormi(x, @inverse(a))
rmvnormic(x, @cholesky(@inverse(a))
Cross-references
For random generator functions, see
“Statistical Distributions” and in particular,
@mrnd,
@mnrnd, and
@rmvnorm.
For related commands, see
nrnd,
rnd,
rndint, and
rmvnorm. See also
rndseed.
Generate uniform random draws.
The rnd command fills series, vector, and matrix objects with (pseudo) random values drawn uniformly from zero to ones. When used with a series, rnd command ignores the current sample and fills the entire object.
Syntax
rnd(object_name)
Fill object_name with uniform random numbers.
Examples
matrix(30, 4) m1
rnd(m1)
Cross-references
For random generator functions, see
“Statistical Distributions” and in particular,
@mrnd,
@mnrnd, and
@rmvnorm.
For related commands, see
nrnd,
rndint, and
rmvnorm. See also
rndseed.
Generate uniform random integers.
The rndint command fills series, vector, and matrix objects with (pseudo) random integers drawn uniformly from zero to a user specified maximum. The rndint command ignores the current sample and fills the entire object with random integers.
Syntax
rndint(object_name, n)
Type the name of the series, vector, or matrix object to fill, followed by an integer value representing the maximum value n of the random integers. n should a positive integer.
Examples
series index
rndint(index,10)
fills the entire series INDEX with integers drawn randomly from 0 to 10. Note that unlike standard series assignment using genr, rndint ignores the current sample and fills the series for the entire workfile range.
sym(3) var3
rndint(var3,5)
fills the entire symmetric matrix VAR3 with random integers ranging from 0 to 5.
Cross-references
For random generator functions, see
“Statistical Distributions” and in particular,
@mrnd,
@mnrnd, and
rmvnorm.
For related commands, see
nrnd,
rnd, and
rmvnorm. See also
rndseed.
Seed the random number generator.
Use rndseed when you wish to generate a repeatable sequence of random numbers, or to select the generator to be used.
Note that EViews 5 has updated the seeding routines of two of our pseudo-random number generators (backward compatible options are provided). It is strongly recommended that you use new generators.
Syntax
rndseed(options) integer
Follow the rndseed keyword with the optional generator type and an integer for the seed.
Options
type=arg (default=“kn”) | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4)”, L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
When EViews starts up, the default generator type is set to the improved Knuth lagged Fibonacci generator. Unless changed using rndseed, Knuth’s generator will be used for subsequent pseudo-random number generation.
| Knuth (“kn4”) | L’Ecuyer (“le”) | Mersenne Twister (“mt4”) |
Period | | | |
Time (for 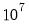 draws) | 27.3 secs | 15.7 secs | 1.76 secs |
Cases failed Diehard test | 0 | 0 | 0 |
Examples
rndseed 123456
genr t3 = @qtdist(rnd,3)
rndseed 123456
genr t30 = @qtdist(rnd,30)
generates random draws from a t-distribution with 3 and 30 degrees of freedom using the same seed.
Cross-references
See the list of available random number generators in
“Statistical Distributions”.
At press time, further information on the improved seeds may be found on the web at the following addresses:
See also
nrnd,
rnd,
rndint, and
rmvnorm.
References
Knuth, D. E. (1997). The Art of Computer Programming, Volume 2, Semi-numerical Algorithms, 3rd edition, Reading, MA: Addison-Wesley Publishing Company. Note: the C implementation of the lagged Fibonacci generator is described in the errata to the 2nd edition, downloadable from Knuth's web site.
L’Ecuyer, P. (1999). “Good Parameters and Implementations for Combined Multiple Recursive Random Number Generators,” Operations Research, 47(1), 159-164
Matsumoto, M. and T. Nishimura (1998). “Mersenne Twister: A 623-Dimensionally Equidistributed Uniform Pseudo-Random Number Generator,” ACM Transactions on Modeling and Computer Simulation, 8(1), 3-30.
Estimates an equation using robust least squares.
You may perform three different types of robust estimation: M-estimation, S-estimation and MM-estimation.
Syntax:
robustls(options) y x1 [x2 x3…]
Enter the robustls keyword, followed by the dependent variable and a list of the regressors.
Options
method=arg (default=“m”) | Robust estimation method: “m” (M-estimation), “s” (S-estimation) or “mm” (MM-estimation). |
cov=arg (default=“type1”) | Covariance method type: “type1”, “type2”, or “type3”. |
tuning=number | Specify a value for the tuning parameter. If a value is not specified, EViews will use the default tuning parameter for the type of estimation and weighting function (if applicable). |
c=s | Convergence criterion. The criterion will be set to the nearest value between 1e-24 and 0.2. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
m=integer | Maximum number the number of iterations. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
M-estimation Options
fn=arg (default=“bisquare”) | Weighting function used during M-estimation: “andrews” (Andrews), “bisquare” (Bisquare), “cauchy” (Cauchy), “fair”, “huber”, “huberbi” (Huber-bisquare), “logistic” (Logistic), “median”, “tal” (Talworth), “Welsch” (Welsch). |
scale=arg (default=“madzero”) | Scaling method used for calculating the scalar parameter during M estimation: “madzero” (median absolute deviation, zero centered), “madmed” (median absolute deviation, median centered), "huber" (Huber scaling). |
hmat | Use the hat-matrix to down-weight observations with high leverage. |
S and MM estimation options
compare = integer (default=4) | Number of comparison sets. |
refine = integer (default= 2) | Number of refinements. |
trials = integer (default=200) | Number of trials. |
subsmpl=integer | Specifies the size of the subsamples. Note, the default is number of coefficients in the regression. |
seed=number | Specifies the random number generator seed |
rng=arg | Specifies the type of random number generator. The key can be; improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple, recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
MM estimation options
mtuning=arg | M-estimator tuning parameter. Note the S-estimator tuning parameter is set with the “tuning=” option outlined above. |
hmat | Use the hat-matrix to down-weight observations with high leverage during m-estimation. |
Examples
The following examples use the “Rousseeuw and Leroy.wf1” file located in the EViews application data directory.
robustls salinity c lagsal trend discharge
This line estimates a simple M-type robust estimation, with SALINITY as the dependent variable, and a constant, LAGSAL, TREND and DISCHARGE as independent variables.
The line:
robustls(method=mm, tuning=2.937, mtuning=3.44, cov=type2) salinity c lagsal trend discharge
estimates the same model, but using MM-estimation, with an S tuning constant of 2.937, an M tuning constant of 3.44, and using Huber Type II standard errors.
Cross-references
Place vector in row of a matrix.
Place a column or rowvector object in a specified row of a matrix.
Syntax
rowplace(m, r, n)
Places the column vector or rowvector r into the matrix m at row n. The number of columns in m and r must match, and row n must exist within m.
Examples
matrix m1 = @mnrnd(30, 5)
matrix r1 = @mnrnd(1, 5)
rowplace(m1, r1, 4)
places R1 in the fourth row of M1.
Cross-references
Run a program.
The run command executes a program. The program may be located in memory or stored in a program file on disk.
Syntax
run(options) [path\]prog_name(prog_options) [%0 %1 …]
If you wish to pass one or more options to the program, you should enclose them in parentheses immediately after the filename. If the program has arguments, you should list them after the filename.
EViews first checks to see if the specified program is in memory. If the program is not located, EViews then looks for the program on disk in the current working directory, or in the specified path. The program file should have a “.PRG” extension, which you need not specify in the prog_name.
Options
integer (default=1) | Set maximum errors allowed before halting the program. |
c | Run program file without opening a window for display of the program file. |
verbose / quiet | Verbose mode in which messages will be sent to the status line at the bottom of the EViews window (slower execution), or quiet mode which suppresses workfile display updates (faster execution). |
v / q | Same as [verbose / quiet]. |
ver4 / ver5 | Execute program in [version 4 / version 5] compatibility mode. |
this=object_name | Set the _this object for the executed program. If omitted, the executed program will inherit the _this object from the parent program, or from the current active workfile object when the exec command is issued from the command window. |
Examples
run(q) simul(h=2) x xhat
quietly runs a program named “Simul.prg” from the default directory using options string “h=2” and arguments %0=X and %1=XHAT.
Since run is a command, it may also be placed in a program file. You should note that if you put the run command in a program file and then execute the program, EViews will stop after executing the program referred to by the run command. For example, if you have a program containing:
run simul
print x
the
print statement will not be executed since execution will stop after executing the commands in “Simul.prg”. If this behavior is not intended, you should consider using the
exec command or an
include statement.
Cross-references
See
“Executing a Program” for further details.
See also
exec and
include.
Save current workfile to disk.
This usage is provided only for backward compatibility, as it has been replaced with the equivalent
wfsave command.
Syntax
save [path\]file_name
Follow the keyword with a name for the file. If an explicit path is not specified, the file will be stored in the default directory, as set in the global options.
Examples
save MyWorkfile
saves the current workfile with the name MYWORKFILE.WF1 in the default directory.
save c:\data\MyWF1
saves the current workfile with the name MYWF1.WF1 in the specified directory.
Cross-references
Saves program variables to an “.ini” file.
Syntax
saveprogini([file=filename]) [section]name value
Saves the value (either string or scalar) into the name under the section [section] in the “.ini” file specified by filename. If no section is provided, a [default] section is used. If no filename is given, EViews will create an “.ini” file with the same name as the current running program in the same directory. If the program is untitled, the current default directory is used. This command must be run from inside a program.
Example
saveprgini(file="c:\temp\myini.ini") hello 3
This will create a “.ini” file containing:
[default]
hello=3
You may then load the value into a string using
@loadprgini:
create u 10
string a = @loadprgini("hello", "c:\temp\myini.ini")
Cross-references
Seasonal adjustment.
The seas command carries out seasonal adjustment using either the ratio to moving average, or the difference from moving average technique.
EViews also performs Census X11 and X12 seasonal adjustment. For details, see
Series::x11 and
Series::x12.
Syntax
seas(options) series_name name_adjust [name_fac]
List the name of the original series and the name to be given to the seasonally adjusted series. You may optionally include an additional name for the seasonal factors. seas will display the seasonal factors using the convention of the Census X11 program.
Options
m | Multiplicative (ratio to moving average) method. |
a | Additive (difference from moving average) method. |
prompt | Force the dialog to appear from within a program. |
Examples
seas(a) pass pass_adj pass_fac
seasonally adjusts the series PASS using the additive method, and saves the adjusted series as PASS_ADJ and the seasonal factors as PASS_FAC.
Cross-references
See
“Seasonal Adjustment” for a discussion of seasonal adjustment methods.
Insert contents into cell of a table.
The setcell command puts a string or number into a cell of a table.
Syntax
setcell(table_name, r, c, content[, "options"])
Options
Provide the following information in parentheses in the following order: the name of the table object, the row number, r, of the cell, the column number, c, of the cell, a number or string to put in the cell, and optionally, a justification and/or numerical format code. A string of text must be enclosed in double quotes.
The justification options are:
c | Center the text/number in the cell. |
r | Right-justify the text/number in cell. |
l | Left-justify the text/number in cell. |
The numerical format code determines the format with which a number in a cell is displayed; cells containing strings will be unaffected. The format code can either be a positive integer, in which case it specifies the number of decimal places to be displayed after the decimal point, or a negative integer, in which case it specifies the total number of characters to be used to display the number. These two cases correspond to the Fixed decimal and Fixed character fields in the number format dialog.
Note that when using a negative format code, one character is always reserved at the start of a number to indicate its sign, and that if the number contains a decimal point, that will also be counted as a character. The remaining characters will be used to display digits. If the number is too large or too small to display in the available space, EViews will attempt to use scientific notation. If there is insufficient space for scientific notation (six characters or less), the cell will contain asterisks to indicate an error.
Examples
setcell(tab1, 2, 1, "Subtotal")
puts the string “Subtotal” in row 2, column 1 of the table object named TAB1.
setcell(tab1, 1, 1, "Price and cost", "r")
puts the a right-justify string “Price and cost” in row 1, column 1 of the table object named TAB1.
Note that in general, that unless you wish to control the formatting, assignment statements of the form
Mytable(1,1) = "hello"
are easier than using the default setcell statement:
Setcell(mytable,1,1,"hello")
Cross-references
“Working with Tables and Spreadsheets” describes table formatting using commands. See
“Table Objects” for a discussion and examples of table formatting in EViews.
See also
Table::setjust and
Table::setformat. Note that this command is supported primarily for backward compatibility. There is a more extensive set of table procs for working with and customizing tables. See
“Table Procs”.
Set width of a column of a table.
Provided for backward compatibility. See
Table::setwidth for the new method of setting the width of table and spreadsheet columns.
Syntax
setcolwidth(table_name, c, width)
Options
To change the width of a column, provide the following information in parentheses, in the following order: the name of the table, the column number c, and the number of characters width for the new width. EViews measures units in terms of the width of a numeric character. Because different characters have different widths, the actual number of characters that will fit may differ slightly from the number you specify. By default, each column is approximately 10 characters wide.
Examples
setcolwidth(mytab,2,20)
sets the second column of table MYTAB to fit approximately 20 characters.
Cross-references
“Working with Tables and Spreadsheets” describes table formatting using commands. See also
“Table Objects” for a discussion and examples of table formatting in EViews.
Note that this command is supported primarily for backward compatibility. There is a more extensive set of table procs for working with and customizing tables. See
“Table Procs”.
Set a user-specified execution error.
Syntax
seterr string
sets an execution error using the specified string. May only be used in programs.
Cross-references
Set the current program execution error count.
Syntax
seterrcount integer
sets the current error count to the specified integer value. May only be used in programs.
Cross-references
Set the maximum number of errors that a program may encounter before execution is halted.
Syntax
setmaxerrs integer
sets the maximum number of errors to the specified integer value.
Cross-references
Place a double horizontal line in a table.
Provided for backward compatibility. For a more general method of setting the line characteristics and borders for a set of table cells, see the table proc
Table::setlines See also
Table::setwidth and
Table::setheight .
Syntax
setline(table_name, r)
Options
Specify the name of the table and the row number r in which to place the horizontal line.
Examples
setline(tab3,8)
places a (double) horizontal line in the eighth row of the table object TAB3.
Cross-references
“Working with Tables and Spreadsheets” describes table formatting using commands. See also
“Table Objects” for a discussion and examples of table formatting in EViews.
See
Table::setlines for more flexible line drawing tools. Note that this command is supported primarily for backward compatibility. There is a more extensive set of table procs for working with and customizing tables. See
“Table Procs”.
Start the Windows command shell, optionally executing a command.
Syntax
shell(options) [arg1 arg2 arg3…]
See
spawn for available options. By default, the Windows command shell will be started in hidden mode with the exit code for success set to zero.
Examples
shell mkdir c:\newdir
makes a new directory “c:\newdir”.
shell(out=flist) dir /b *.wf1
lists all workfiles in the current directory, saving output in a table named FLIST.
Cross-references
See
spawn for details on spawning a new process.
Display objects.
The show command displays series or other objects on your screen. A scalar object is displayed in the status line at the bottom of the EViews window.
Syntax
show object_name.view_command
show object1 [object2 object3 ...]
The command show should be followed by the name of an object, or an object name with an attached view.
For series and graph objects, show can operate on a list of names. The list of names must be of the same type. show creates and displays an untitled group or multiple graph object.
Examples
genr x=nrnd
show x.hist
close x
generates a series X of random draws from a standard normal distribution, displays the histogram view of X, and closes the series window.
show wage log(wage)
opens an untitled group window with the spreadsheet view of the two series.
freeze(gra1) wage.hist
genr lwage=log(wage)
freeze(gra2) lwage.hist
show gra1 gra2
opens an untitled graph object with two histograms.
Cross-references
See
“Object Commands” for discussion.
“Object View and Procedure Reference” provides a complete listing of the views of the various objects.
Forecasts a series using one of a number of exponential smoothing techniques. By default, smooth estimates the damping parameters of the smoothing model to minimize the sum of squared forecast errors, but you may specify your own values for the damping parameters.
smooth automatically calculates in-sample forecast errors and puts them into the series RESID.
Syntax
smooth(method) series_name smooth_name [freq]
You should follow the smooth keyword with the name of the series to smooth and a name for the smoothed series. You must also specify the smoothing method in parentheses. The optional freq may be used to override the default for the number of periods in the seasonal cycle. By default, this value is set to the workfile frequency (e.g. — 4 for quarterly data). For undated data, the default is 5.
Options
Smoothing method options
s[,x] | Single exponential smoothing for series with no trend. You may optionally specify a number x between zero and one for the mean parameter. |
d[,x] | Double exponential smoothing for series with a trend. You may optionally specify a number x between zero and one for the mean parameter. |
n[,x,y] (default) | Holt-Winters without seasonal component. You may optionally specify numbers x and y between zero and one for the mean and trend parameters, respectively. |
a[,x,y,z] | Holt-Winters with additive seasonal component. You may optionally specify numbers x, y, and z, between zero and one for the mean, trend, and seasonal parameters, respectively. |
m[,x,y,z] | Holt-Winters with multiplicative seasonal component. You may optionally specify numbers x, y, and z, between zero and one for the mean, trend, and seasonal parameters, respectively. |
Other Options:
forcsmpl = arg | Forecast sample (optional). If forecast sample is not provided, the workfile sample will be employed. |
prompt | Force the dialog to appear from within a program. |
p | Print a table of forecast statistics. |
If you wish to set only some of the damping parameters and let EViews estimate the other parameters, enter the letter “e” where you wish the parameter to be estimated.
If the number of seasons is different from the frequency of the workfile (an unusual case that arises primarily if you are using an undated workfile for data that are not monthly or quarterly), you should enter the number of seasons after the smoothed series name. This optional input will have no effect on forecasts without seasonal components.
Examples
smooth(s) sales sales_f
smooths the SALES series by a single exponential smoothing method and saves the smoothed series as SALES_F. EViews estimates the damping (smoothing) parameter and displays it with other forecast statistics in the SALES series window.
smooth(n,e,.3) tb3 tb3_hw
smooths the TB3 series by a Holt-Winters no seasonal method and saves the smoothed series as TB3_HW. The mean damping parameter is estimated while the trend damping parameter is set to 0.3.
smpl @first @last-10
smooth(m,.1,.1,.1) order order_hw
smpl @all
graph gra1.line order order_hw
show gra1
smooths the ORDER series by a Holt-Winters multiplicative seasonal method leaving the last 10 observations. The damping parameters are all set to 0.1. The last three lines plot and display the actual and smoothed series over the full sample.
Cross-references
See
“Exponential Smoothing” for a discussion of exponential smoothing methods.
Set sample range.
The smpl command sets the workfile sample to use for statistical operations and series assignment expressions.
Syntax
smpl smpl_spec
smpl sample_name
List the date or number of the first observation and the date or number of the last observation for the sample. Rules for specifying dates are given in
“Dates”. The sample spec may contain more than one pair of beginning and ending observations.
The smpl command also allows you to select observations on the basis of conditions specified in an if statement. This enables you to use logical operators to specify what observations to include in EViews’ procedures. Put the if statement after the pairs of dates.
You can also use smpl to set the current observations to the contents of a named sample object; put the name of the sample object after the keyword.
Special keywords for smpl
The following “@-keywords” can be used in a smpl command:
@all | The entire workfile range. |
@first | The first observation in the workfile. |
@last | The last observation in the workfile. |
The following frequency functions may also be used:
@year | The four digit year in which the current observation begins. |
@quarter | The quarter of the year in which the current observation begins. |
@month | The month of the year in which the current observation begins. |
@day | The day of the month in which the current observation begins. |
@weekday | The day of the week in which the current observation begins, where Monday is given the number 1 and Sunday is given the number 7. |
@hour | The observation hour as an integer. (9:30AM returns 9, 5:15PM returns 17.) |
@minute | The observation minute as an integer. (9:30PM returns 30.) |
@second | The observation second as an integer. |
@hourf | The observation time as a floating point hour. (9:30AM returns 9.5, 5:15PM returns 17.25.) |
In panel settings, you may use the additional keywords:
@firstmin | The earliest of the first observations (computed across cross-sections). |
@firstmax | The latest of the first observations. |
@lastmin | The earliest of the last observations. |
@lastmax | The latest of the last observations. |
Examples
smpl 1955m1 1972m12
sets the workfile sample from 1955M1 to 1972M12.
smpl @first 1940 1946 1972 1975 @last
excludes observations (or years) 1941–1945 and 1973–1974 from the workfile sample.
smpl if union=1 and edu<=15
sets the sample to those observations where UNION takes the value 1 and EDU is less than or equal to 15.
sample half @first @first+@obs(x)/2
smpl half
smpl if x>0
smpl @all if x>0
The first line declares a sample object named HALF which includes the first half of the series X. The second line sets the sample to HALF and the third line sets the sample to those observations in HALF where X is positive. The last line sets the sample to those observations where X is positive over the full sample.
The sample may be set for intraday data using optional times after the dates. For example,
smpl 1/3/2000 10AM 12/30/2000 2PM
removes any observations before 10AM on 1/3/2000 and after 2PM on 12/30/2000.
smpl if @hourf<=9.5 and @hourf<=14.5
sets the sample to include only observations between and including 9:30AM and 2:30PM.
smpl if @minute=0 or @minute=30
selects only observations that appear on the half hour.
smpl if @weekday=1 and @hourf=10
sets the sample to include only observations that appear on Mondays at 10AM.
Cross-references
See
“Samples” for a discussion of samples in EViews.
Solve the model.
solve finds the solution to a simultaneous equation model for the set of observations specified in the current workfile sample.
Syntax
solve(options) model_name
Note: when solve is used in a program (batch mode) models are always solved over the workfile sample. If the model contains a solution sample, it will be ignored in favor of the workfile sample.
You should follow the name of the model after the solve command. The default solution method is dynamic simulation. You may modify the solution method as an option.
solve first looks for the specified model in the current workfile. If it is not present, solve attempts to fetch a model file (.DBL) from the default directory or, if provided, the path specified with the model name.
Options
solve can take any of the options available in
Model::solveopt.
Examples
solve mod1
solves the model MOD1 using the default solution method.
solve(m=500,e) nonlin2
solves the model NONLIN2 with an extended search of up to 500 iterations.
Cross-references
See
“Models” for a discussion of models.
Sort the current workfile page.
The sort command sorts all series in the workfile page on the basis of the values of one or more of the series. For purposes of sorting, NAs are considered to be smaller than any other value. By default, EViews will sort the series in ascending order. You may use options to override the sort order.
EViews will first remove any workfile structures and then will sort the workfile using the specified settings.
Syntax
sort(options) arg1 [arg2 arg3…]
List the name of the series or groups by which you wish to sort the workfile. If you list two or more series, sort uses the values of the second series to resolve ties from the first series, and values of the third series to resolve ties from the second, and so on.
Options
d | sort in descending order. |
Examples
sort(d) inc
sorts all series in the workfile in order of the INC series with the highest value of INC first. NAs in INC (if any) will be placed at the bottom.
sort gender race wage
sorts all series in the workfile in order of the values of GENDER from low to high, with ties resolved by ordering on the basis of RACE, with further ties resolved by ordering on the basis of WAGE.
Cross-references
Spawn a new process.
Syntax
spawn(options) filename [arg1 arg2 arg3…]
Follow the keyword with a filename indicating the process to spawn, and optional arguments to be passed to the process.
Options
“n” or “normal” | Create process in normal mode. The process will typically create a maximized window and may wait for user input. |
“m” or “minimized” | Create process in a minimized window. Note that some applications may not accept requests to run in minimized mode. |
“h” or “hidden” | Create process in hidden mode (without a visible window). Note that some applications may not accept requests to run in hidden mode. |
t=isecs | Specifies the maximum time in milliseconds that EViews should wait for the process to complete. If the timeout interval is reached and the process has not completed, EViews will generate an error. A timeout setting of zero may be used to indicate that EViews should not wait for the spawned process to complete. If no timeout option is provided, EViews will wait indefinitely for the process to complete. |
exit=icode | Specifies the exit code that the process will return if it is completed successfully. If the process returns an exit code other than the specified value, EViews will generate an error. If the exit code option is not specified, EViews will not generate an error no matter what exit code is returned by the process. |
out = tablename | If an output table name is specified, EViews will capture any data written to standard output by the spawned process and store it into a table object with the specified name in the workfile. Note that this option will have no effect unless either the minimized or hidden option is used and the timeout value is not zero. |
Examples
spawn "c:\program files\microsoft office\office11\excel.exe" test.xls
starts a new Excel process, passing it the command line argument “test.xls”.
Cross-references
See
shell for information on starting a Windows command shell.
Descriptive statistics.
Computes and displays a table of means, medians, maximum and minimum values, standard deviations, and other descriptive statistics of one or more series or a group of series.
stats creates an untitled group containing all of the specified series, and opens a statistics view of the group. By default, if more than one series is given, the statistics are calculated for the common sample.
Syntax
stats(options) ser1 [ser2 ser3 …]
Options
Examples
stats height weight age
opens an untitled group window displaying the histogram and descriptive statistics for the common sample of the three series.
Cross-references
See
“Descriptive Statistics & Tests” and
(here) for a discussion of the descriptive statistics views of series and groups.
See also
boxplot and
hist.
Send text to the status line.
Displays a message in the status line at the bottom of the EViews main window. The message may include text, control variables, and string variables.
Syntax
statusline message_string
Examples
statusline Iteration Number: !t
Displays the message “Iteration Number: !t” in the status line replacing “!t” with the current value of the control variable in the program.
Cross-references
See
“EViews Programming” for a discussion and examples of programs, control variables and string variables.
Convert series or group to a matrix object after removing NAs.
eries-TO-atrix object: convert the data in a series or group or series into a matrix after removing rows with missing values.
Syntax
stom(o1, o2, smp)
where o1 is a series, alpha, or group, o2 is a corresponding vector, svector, or matrix, and smp is an optional sample.
• If o1 is a series, stom fills the vector o2 with data from the o1 using the optional sample object smp or the workfile sample. o2 will be resized accordingly. If any observation has the value “NA”, the observation will be omitted from the vector.
• If o1 is a group, stom fills the matrix o2 with data from o1 using the optional sample object smp or the workfile sample. o2 will be resized accordingly. The series in o1 are placed in the columns of o2 in the order they appear in the group spreadsheet. If any of the series in the group has the value “NA” for a given series observation or blank for an alpha, the observation will be omitted for all series.
• If o1 is an alpha-series, stom fills the svector o2 with data from o1 using the optional sample object smp or the workfile sample. o2 will be resized accordingly. If any of the observation is blank, the observation will be omitted from the result.
For a conversion method that preserves NAs, see
stomna.
Examples
stom(ser1,v1)
stom(ser1,v2,smp1)
stom(grp1,m1)
stom(grp1,m2,smp1)
Cross-references
Convert series or group to a matrix object, retaining NAs.
eries-TO-atrix object with s: convert the data in a series or group or series into a matrix, retaining rows with missing values.
Syntax
stom(o1, o2, smp)
where o1 is a series, alpha, or group, o2 is a corresponding vector, svector, or matrix, and smp is an optional sample.
• If o1 is a series, stomna fills the vector o2 with data from o1 using the optional sample object smp or the workfile sample. o2 will be resized accordingly. All “NA” values in the series will be assigned to the corresponding vector elements.
• If o1 is a group, stomna fills the matrix o2 with data from o1 using the optional sample object smp or the workfile sample. o2 will be resized accordingly. The series in o1 are placed in the columns of o2 in the order they appear in the group spreadsheet. All NAs will be assigned to the corresponding matrix elements.
• If o1 is an alpha-series, stomna fills the svector o2 with data from o1 using the optional sample object smp or the workfile sample. o2 will be resized accordingly. All “NA” values (blank elements of the alpha) will be assigned to the corresponding vector elements.
For conversion methods that automatically remove observations with NAs, see
@convert and
stom.
Examples
stom(ser1,v1)
stom(ser1,v2,smp1)
stomna(grp1,m1)
stomna(grp1,m2,smp1)
Cross-references
See also
mtos,
@convert,
stom, and
ttom.
Store objects in databases and databank files.
Stores one or more objects in the current workfile in EViews databases or individual databank files on disk. The objects are stored under the name that appears in the workfile.
Syntax
store(options) object_list
Follow the store command keyword with a list of object names (each separated by a space) that you wish to store. The default is to store the objects in the default database. (This behavior is a change from EViews 2 and earlier where the default was to store objects in individual databank files).
You may precede the object name with a database name and the double colon “::” to indicate a specific database. You can also specify the database name as an option in parentheses, in which case all objects without an explicit database name will be stored in the specified database.
You may use wild card characters “?” (to match any single character) or “*” (to match zero or more characters) in the object name list. All objects with names matching the pattern will be stored.
You can optionally choose to store the listed objects in individual databank files. To store in files other than the default path, you should include a path designation before the object name.
Options
d=db_name | Store to the specified database. |
i | Store to individual databank files. |
1 / 2 | Store series in [single / double] precision to save space. |
o | Overwrite object in database (default is to merge data, where possible). |
g=arg | Group store from workfile to database: “s” (copy group definition and series as separate objects), “t” (copy group definition and series as one object), “d” (copy series only as separate objects), “l” (copy group definition only). |
If you do not specify the precision option (1 or 2), the global option setting will be used. See
“Database Storage Defaults”.
Examples
store m1 gdp unemp
stores the three objects M1, GDP, UNEMP in the default database.
store(d=us1) m1 gdp macro::unemp
stores M1 and GDP in the US1 database and UNEMP in the MACRO database.
store usdat::gdp macro::gdp
stores the same object GDP in two different databases USDAT and MACRO.
store(1) cons*
stores all objects with names starting with CONS in the default database. The “1” option uses single precision to save space.
store(i) m1 c:\data\unemp
stores M1 and UNEMP in individual databank files.
Cross-references
“Basic Data Handling” discusses exporting data in other file formats. See
“EViews Databases” for a discussion of EViews databases and databank files.
For additional discussion of wildcards, see
Appendix A. “Wildcards”.
Estimate a switching regression model (simple exogenous or Markov).
Syntax
switchreg(options) dependent_var list_of_varying_regressors [ @nv list_of_nonvarying_regressors ] [ @prv list_of_probability_regressors ]
List the switchreg keyword, followed by options, then the dependent variable and a list of the regressors with regime-varying coefficients, following optionally by the keyword @nv and a list of regressors with regime-invariant coefficients, and by the keyword @prv and a list of regressors that enter into the transition probability specification.
The dependent variable in switchreg may not be an expression. Dynamics may be specified by including lags of the dependent variable as regressors, or by specifying AR errors using the AR keyword. The latter incorporate mean adjusted lags of the form specified by the “Hamilton-model.”
Options
type=arg | Type of switching: simple exogenous (“simple”), Markov (“markov”). |
nstates=integer (default=2) | Number of regimes. |
heterr | Allow for heterogeneous error variances across regimes |
fprobmat=arg | Name of fixed transition probability matrix allows for fixing specific elements of the time-invariant transition matrix. Leave NAs in elements of the matrix to estimate. The 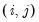 element of the matrix corresponds to  . |
initprob=arg (default=“ergodic”) | Method for determining initial Markov regime probabilities: ergodic solution (“ergodic”), estimated parameter (“est”), equal probabilities (“uniform”), user-specified probabilities (“user”). If “initprob=user” is specified, you will need to specify the “userinit=” option. |
userinit=arg | Name of vector containing user-specified initial Markov probabilities. The vector should have rows equal to the number of states; we expand this to the size of the initial lag state vector where necessary for AR specifications. For use in specifications containing both the “type=markov” and “initprob=user” options. |
startnum=arg (default=0 or 25) | Number of random starting values tried. The default is 0 for user-supplied coefficients (option “s”) and 25 in all other cases. |
startiter=arg (default=10) | Number of iterations taken after each random start before comparing objective to determine final starting value. |
searchnum=arg (default=0) | Number of post-estimation perturbed starting values tried. |
searchstds=arg (default=1) | Number of standard deviations to use in perturbed starts (if “searchnum=”) is specified. |
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
rnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
In addition to the specification options, there are options for estimation and covariance calculation.
Additional Options
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). BFGS is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
switchreg(type=markov) y c @nv ar(1) ar(2) ar(3) ar(4)
estimates a Hamilton-type Markov switching regression model with four non-regime varying autoregressive terms implying mean adjustment for the lagged endogenous.
switchreg(type=markov) y c @nv y(-1) y(-2) y(-3) y(-4)
specifies an alternate dynamic model in which the lags enter directly into the contemporaneous equation without mean adjustment.
switchreg(type=markov) yy_dalt c @nv ar(1) ar(2) ar(3) ar(4) @prv c yy_ldalt
estimates a 2 state model with non-varying AR(4) and transition matrix probability regressor YY_LDALT.
Cross-references
See
“Switching Regression” for a description of the switching regression methodology.
See also
Equation::rgmprobs,
Equation::transprobs,
Equation::makergmprobs and
Equation::maketransprobs for routines that allow you to work with the regime probabilities and transition probabilities.
Copies a portion of one table to the specified location in another table.
Syntax
tabplace(desttable,sourcetable,d1,s1,s2)
tabplace(desttable,sourcetable,dr1,dc1,sr1,sc1,sr2,sc2)
The tabplace command can be specified either using coordinates where columns are signified with a letter, and rows by a number (for example “A3” represents the first column, third row), or by row number and column number.
The first syntax represents coordinate form, where sourcetable is the name of the table from which to copy, s1 specifies the upper-left coordinate portion of the section of the source table to be copied, s2 specifies the bottom-right coordinate, desttable specifies the name of the table to copy to, and d1 specifies the upper-left coordinate of the destination table.
The second syntax represents the row/column number form, where sourcetable is the name of the table from which to copy, sr1 specifies the source table upper row number, sc1 specifies the source table left most column number, sr2 specifies the source table bottom row number, sc2 specifies the source table right most column number. desttable specifies the name of the table to copy to, and dr1 and dr2 specify the upper and left most row and column of the destination table, respectively.
Examples
tabplace(table2,table1,"d1","B9","E17")
places a copy of the data from cell range B9 to E17 in TABLE1 to TABLE2 at cell D1
tabplace(table3,table1,10,3,9,2,17,5)
copies 8 rows of data (from row 9 to row 17) and 3 columns (from 2 to 5)of data in TABLE1 to the tenth row and 3rd column of TABLE3.
Cross-references
For additional discussion of table commands see
“Working with Tables and Spreadsheets”.
See also
“Table and Text Objects” for a discussion and examples of table formatting in EViews.
Test whether to add regressors to an estimated equation.
Tests the hypothesis that the listed variables were incorrectly omitted from an estimated equation (only available for equations estimated by list). The test displays some combination of Wald and LR test statistics, as well as the auxiliary regression.
Syntax
testadd(options) arg1 [arg2 arg3 ...]
List the names of the series or groups of series to test for omission after the keyword. The test is applied to the default equation, if defined.
Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
ls sales c adver lsales ar(1)
testadd gdp gdp(-1)
tests whether GDP and GDP(-1) belong in the specification for SALES. The commands:
Cross-references
See
“Coefficient Diagnostics” for further discussion.
Test whether to drop regressors from a regression.
Tests the hypothesis that the listed variables were incorrectly included in the estimated equation (only available for equations estimated by list). The test displays some combination of
and LR test statistics, as well as the test regression.
Syntax
testdrop(options) arg1 [arg2 arg3 ...]
List the names of the series or groups of series to test for omission after the keyword. The test is applied to the default equation, if defined.
Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
ls sales c adver lsales ar(1)
testdrop adver
tests whether ADVER should be excluded from the specification for SALES. The commands:
Cross-references
See
“Coefficient Diagnostics” for further discussion of testing coefficients.
Estimation by discrete or smooth threshold least squares, including threshold autoregression.
Syntax
threshold(options) y z1 [z2 z3 ...] [@nv x1 x2 x3 ...] @thresh t1 [t2 t3 ...]
List the dependent variable first, followed by a list of the independent variables that have coefficients that are allowed to vary across thresholds, followed optionally by the keyword @nv and a list of non-varying coefficient variables.
List a threshold variable or variables (for model selection) or a single integer or range pairs after the keyword @thresh. The integer or range pairs indicate a self-exciting model with the lagged dependent variable as the threshold variable.
For smooth threshold equations you may specify variables that are to be included only in the base specification or only in the alternative specification. Base-only variables should be specified in parentheses using the @base key, as in “@base(x1) @base(x2) @base(x3 x4)”. Alternative-only variables may be specified analogously using the @alt key.
Options
Specification Options
type=arg (default=“discrete”) | Type of threshold estimation: “discrete” (discrete), “smooth” (smooth). |
Discrete Threshold Options
method=arg (default=“seqplus1”) | Threshold selection method: “seqplus1” (sequential tests of single  versus  thresholds), “seqall” (sequential test of all possible 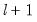 versus 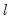 thresholds), “glob” (tests of global 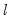 vs. no thresholds), “globplus1” (tests of  versus  globally determined thresholds), “globinfo” (information criteria evaluation)., “fixedseq” (fixed number of sequentially determined thresholds), “fixedglob” (fixed number of globally determined thresholds), “user” (user-specified thresholds) |
nthresh=arg (default=1) | Number of thresholds for fixed number threshold selection methods. |
select=arg | Sub-method setting (options depend on “method=”). (1) if “method=glob”: Sequential ("seq") (default), Highest significant ("high"), 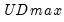 ("udmax"), 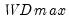 ("wdmax"). (2) if “method=globinfo”: Schwarz criterion (“bic” or “sic”) (default), Liu-Wu-Zidek criterion (“lwz”). |
trim=arg (default=5) | Trimming percentage for determining minimum segment size (5, 10, 15, 20, 25). |
maxthresh=integer (default=5) | Maximum number of thresholds to allow (not applicable if “method=seqall”). |
maxlevels=integer (default=5) | Maximum number of threshold levels to consider in sequential testing (applicable when “method=sequall”). |
size=arg (default=5) | Test sizes for use in sequential determination and final test evaluation (10, 5, 2.5, 1) corresponding to 0.10, 0.05, 0.025, 0.01, respectively |
heterr | Assume regimes specific error distributions in variance computation. |
commondata | Assume a common distribution for the data across segments (only applicable if original equation is estimated with a robust covariance method, “heterr” is not specified). |
Smooth Threshold Options
smoothtrans=arg (default=“logistic”) | Smooth threshold transition function: “logistic” (logistic), “logistic2” (second-order logistic), “exponential” (exponential), “normal” (normal). |
smoothstart=arg (default=“grid_conc”) | Smoth threshold starting value method: or fixed number threshold selection methods: “grid_conc” (grid search with concentrated regression coefficients”, “grid_zeros” (grid search with zero regression coefficients), “data” (data-based), “user” (user-specified using the contents of the coefficient vector in the workfile). |
smoothst=arg | Sub-method setting (options depend on “method=”). (1) if “method=glob”: Sequential ("seq") (default), Highest significant ("high"), 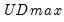 ("udmax"), 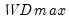 ("wdmax"). (2) if “method=globinfo”: Schwarz criterion (“bic” or “sic”) (default), Liu-Wu-Zidek criterion (“lwz”). |
General Options
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=keyword | Covariance type (optional): “white” (White diagonal matrix), “hac” (Newey-West HAC). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of  . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
threshold(method=fixedseq, type=discrete) ss_transf c ss_transf(-1 to -11) @thresh 2
uses the fixed number of thresholds test to determine the optimal threshold in a model regressing SS_TRANSF on the threshold variables C and SS_TRANSF(-1 to -11).
threshold(method=fixedseq, type=discrete) ss_transf c ss_transf(-1 to -11) @thresh 1 5
uses the fixed number of thresholds test to determine the optimal threshold and does model selection over lags of SS_TRANSF from SS_TRANSF(-1) to SS_TRANSF(-5).
threshold(method=user, threshold=7.44) ss_transf c @nv ss_transf(-1 to -11) @thresh 2
estimates the model with one user-specified threshold value. In addition, the variables SS_TRANSF(-1 to -11) are restricted to have common coefficients across the regimes.
Cross-references
See
“Discrete Threshold Regression” for a discussion of the various forms of threshold models.
Reset the timer.
Syntax
Command: tic
Examples
The sequence of commands:
tic
[some commands]
toc
resets the timer, executes commands, and then displays the elapsed time in the status line. Alternatively:
tic
[some commands]
!elapsed = @toc
resets the time, executes commands, and saves the elapsed time in the control variable !ELAPSED.
Cross-references
Display elapsed time (since timer reset) in seconds.
Syntax
Command: toc
Examples
The sequence of commands:
tic
[some commands]
toc
resets the timer, executes commands, and then displays the elapsed time in the status line. The set of commands:
tic
[some commands]
!elapsed = @toc
[more commands]
toc
resets the time, executes commands, saves the elapsed time in the control variable !ELAPSED, executes additional commands, and displays the total elapsed time in the status line.
Cross-references
See also
tic,
toc, and
@toc.
Two-stage least squares.
Carries out estimation using two-stage least squares.
Syntax
tsls(options) y x1 [x2 x3 ...] @ z1 [z2 z3 ...]
tsls(options) specification @ z1 [z2 z3 ...]
To use the tsls command, list the dependent variable first, followed by the regressors, then any AR or MA error specifications, then an “@”-sign, and finally, a list of exogenous instruments. You may estimate nonlinear equations or equations specified with formulas by first providing a specification, then listing the instrumental variables after an “@”-sign.
There must be at least as many instrumental variables as there are independent variables. All exogenous variables included in the regressor list should also be included in the instrument list. A constant is included in the list of instrumental variables even if not explicitly specified.
Options
Non-Panel TSLS Options
nocinst | Do not automatically include a constant as an instrument. |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
z | Turn off backcasting in ARMA models. |
cov=keyword | Covariance type (optional): “white” (White diagonal matrix), “hac” (Newey-West HAC). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 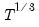 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR or MA terms (see also
param). |
s=number | Determine starting values for equations specified by list with AR or MA terms. Specify a number between zero and one representing the fraction of TSLS estimates computed without AR or MA terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR and MA terms which are set to EViews determined default values. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Panel TSLS Options
cx=arg | Cross-section effects. For fixed effects estimation, use “cx=f”; for random effects estimation, use “cx=r”. |
per=arg | Period effects. For fixed effects estimation, use “cx=f”; for random effects estimation, use “cx=r”. |
wgt=arg | GLS weighting: (default) none, cross-section system weights (“wgt=cxsur”), period system weights (“wgt=persur”), cross-section diagonal weighs (“wgt=cxdiag”), period diagonal weights (“wgt=perdiag”). |
cov=arg | Coefficient covariance method: (default) ordinary, White cross-section system robust (“cov=cxwhite”), White period system robust (“cov=perwhite”), White heteroskedasticity robust (“cov=stackedwhite”), Cross-section system robust/PCSE (“cov=cxsur”), Period system robust/PCSE (“cov=persur”), Cross-section heteroskedasticity robust/PCSE (“cov=cxdiag”), Period heteroskedasticity robust (“cov=perdiag”). |
keepwgts | Keep full set of GLS weights used in estimation with object, if applicable (by default, only small memory weights are saved). |
rancalc=arg (default=“sa”) | Random component method: Swamy-Arora (“rancalc=sa”), Wansbeek-Kapteyn (“rancalc=wk”), Wallace-Hussain (“rancalc=wh”). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
iter=arg (default=“onec”) | Iteration control for GLS specifications: perform one weight iteration, then iterate coefficients to convergence (“iter=onec”), iterate weights and coefficients simultaneously to convergence (“iter=sim”), iterate weights and coefficients sequentially to convergence (“iter=seq”), perform one weight iteration, then one coefficient step (“iter=oneb”). Note that random effects models currently do not permit weight iteration to convergence. |
unbalsur | Compute SUR factorization in unbalanced data using the subset of available observations for a cluster. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default is to use the “C” coefficient vector. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR terms (see also
param). |
s=number | Determine starting values for equations specified by list with AR terms. Specify a number between zero and one representing the fraction of TSLS estimates computed without AR terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR terms which are instead set to EViews determined default values. |
m=integer | Set maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
tsls y_d c cpi inc ar(1) @ lw(-1 to -3)
estimates an UNTITLED equation using TSLS regression of Y_D on a constant, CPI, INC with AR(1) using a constant, LW(-1), LW(-2), and LW(-3) as instruments.
param c(1) .1 c(2) .1
tsls(s,m=500) y_d=c(1)+inc^c(2) @ cpi
estimates a nonlinear TSLS model using a constant and CPI as instruments. The first line sets the starting values for the nonlinear iteration algorithm.
Cross-references
See
“Additional Regression Tools” and
“Two-Stage Least Squares” for details on two-stage least squares estimation in single equations and systems, respectively.
“Instrumental Variables” discusses estimation using pool objects, while
“Instrumental Variables Estimation” discusses estimation in panel structured workfiles.
Convert table to matrix object.
able--atrix Object: fills the matrix, m1, with the numeric values contained in the table o1. m1 will be resized appropriately. o1 may be a table object in the current workfile, or an object view command that creates a table view from an object.
Syntax
ttom(o1, o2[, range="cell_range", keepna])
where o1 is a table object in the current workfile, or an object view command that creates a table view from an object, o2 is a corresponding vector, svector, or matrix, range is an optional range of cells, and a non-zero value for keepna may be used to indicate that missing values should be retained. By default any row or column of the table that does not contain numbers will be dropped from the matrix.
If the “range=” option is provided, a subset of the cells of the table will be converted, where cell_range can take one of the following forms:
@all | Apply to all cells in the table. |
cell | Cell identifier. You can identify cells using either the column letter and row number (e.g., “A1”), or by using “R” followed by the row number followed by “C” and the column number (e.g., “R1C2”). |
row[,] col | Row number, followed by column letter or number (e.g., “2,C”, or “2,3”), separated by “,”. Apply to cell. |
row | Row number (e.g., “2”). Apply to all cells in the row. |
col | Column letter (e.g., “B”). Apply to all cells in the column. |
first_cell[:]last_cell, first_cell[,]last_cell | Top left cell of the selection range (specified in “cell” format), followed by bottom right cell of the selection range (specified in “cell” format), separated by a “:” or “,” (e.g., “A2:C10”, “A2,C10”, or “R2C1:R10C3”, “R2C1,R10C3”). Apply to all cells in the rectangular region defined by the first cell and last cell. |
first_cell_row[,] first_cell_col[,] last_cell_row[,] last_cell_col | Top left cell of the selection range (specified in “row[,] col” format), followed by bottom right cell of the selection range (specified in “row[,] col” format), separated by a “,” (e.g., “2,A,10,C” or “2,1,10,3”). Apply to all cells in the rectangular region defined by the first cell and last cell. |
Examples
ttom(table1,matrix1)
Creates a matrix object, MATRIX1, containing the numerical data in the table object TABLE1. Any row or column of the table that contains no numbers (i.e. are blank rows/columns, or only contain text) are skipped in the conversion process.
ttom(table1, matrix1, keepna)
Performs the same conversion, but this time any row or column of the table that contains no numbers are included in the conversion process as a row/column of NAs.
ttom(eq01.wald c(1)=0.3, matrix1)
Performs a Wald test of the restriction C(1)=0.3 from the EQ01, and saves the numerical output from that test in to the matrix object MATRIX1.
Cross-references
Break links and auto-updating series (formulae) in the specified series objects.
Syntax
unlink link_names
unlink converts link objects and auto-updating series to ordinary series or alphas. Follow the keyword with a list of names of links and auto-updating series to be converted to ordinary series (values). The list of links may include wildcard characters.
Examples
unlink gdp income
converts the link series GDP and INCOME to ordinary series.
unlink *
breaks all links in the active workfile page.
Cross-references
See
“Series Links” for a description of link objects, and
“Auto-Updating Series” for a discussion of auto-updating series. See also
Link::link and
Link::linkto.
See also
pageunlink and
wfunlink for page and workfile based unlinking, respectively.
Carries out unit root tests on a series or panel structured series.
For ordinary series, computes conventional Augmented Dickey-Fuller (ADF), GLS detrended Dickey-Fuller (DFGLS), Phillips-Perron (PP), Kwiatkowski, et. al. (KPSS), Elliot, Rothenberg, and Stock (ERS) Point Optimal, or Ng and Perron (NP) tests for a unit root in the series or its first or second difference.
For series in a panel structured workfile, computes Levin, Lin and Chu (LLC), Breitung, Im, Pesaran, and Shin (IPS), Fisher - ADF, Fisher - PP, or Hadri panel unit root tests on levels, first, or second differences of the data.
Syntax
uroot(options) series_name
There are different options for conventional tests on an ordinary series and panel tests for series in panel structured workfiles.
Options for Conventional Unit Root Tests
Basic Specification
You should specify the exogenous variables and order of dependent variable differencing in the test equation using the following options:
exog=arg (default=“const”) | Specification of exogenous trend variables in the test equation: “const” “trend” (include a constant and a linear time trend), “none” (do not include any exogenous regressors). |
dif=integer (default=0) | Order of differencing of the series prior to running the test. Valid values are {0, 1, 2}. |
You should specify the test type using one of the following keywords:
adf (default) | Augmented Dickey-Fuller. |
dfgls | GLS detrended Dickey-Fuller (Elliot, Rothenberg, and Stock). |
pp | Phillips-Perron. |
kpss | Kwiatkowski, Phillips, Schmidt, and Shin. |
ers | Elliot, Rothenberg, and Stock (Point Optimal). |
np | Ng and Perron. |
Note that for backward compatibility, EViews supports older forms of the exogenous specification:
const, c (default) | Include a constant in the test equation. |
trend, t | Include a constant and a linear time trend in the test equation. |
none, n | Do not include a constant or time trend (only available for the ADF and PP tests). |
For future compatibility we recommend that you use the “exog=” format.
Spectral Estimation Option
In addition, PP, KPSS, ERS, and NP tests all require the estimation of the long-run variance (frequency zero spectrum). You may specify the method using the “hac=” option. The default setting depends on the selected test.
hac=arg (default=varies) | Method of estimating the frequency zero spectrum: “bt” (Bartlett kernel), “pr” (Parzen kernel), “qs” (Quadratic Spectral kernel), “ar” (AR spectral), “ardt (AR spectral - OLS detrended data), “argls” (AR spectral - GLS detrended data). The default settings are test specific (“bt” for PP and KPSS tests, “ar” for ERS, “argls” for NP). |
Lag Difference Options
Applicable to ADF and DFGLS tests, and for PP, KPSS, ERS, and NP tests that use a AR spectral density estimator (“hac=ar”, “hac=ardt”, or “hac=argls”). The default lag selection method is based on a comparison of Schwarz criterion values. You may specify a fixed lag using the “lag=” option.
lagmethod=arg (default=“sic”) | Method for selecting lag length (number of first difference terms) to be included in the Dickey-Fuller test regression or number of lags in the AR spectral density estimator: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “msaic” (Modified Akaike), “msic” (Modified Schwarz), “mhqc” (Modified Hannan-Quinn), “tstat” (Ng-Perron first backward significant t-statistic). |
lag=integer | Use-specified fixed lag. |
maxlag=integer | Maximum lag length to consider when performing automatic lag length selection. default= 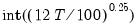 |
lagpval=arg (default=0.1) | Probability value for use in the t-statistic automatic lag selection method (“lagmethod = tstat”). |
Kernel Option
Applicable to PP, KPSS, ERS, and NP tests when using kernel estimators of the frequency zero spectrum (where “hac=bt”, “hac=pz”, or “hac=qs”)
band = arg, b=arg (default=“nw”) | Method of selecting the bandwidth: “nw” (Newey-West automatic variable bandwidth selection), “a” (Andrews automatic selection), number (user specified bandwidth). |
General Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Options for Panel Unit Root Tests
Basic Specification
You should specify the exogenous variables, order of dependent variable differencing, and sample handling, in the test equation using the following options:
exog=arg (default=“const”) | Specification of exogenous trend variables in the test equation: “const” (constant),“trend” (include a constant and a linear time trend), “none” (do not include exogenous regressors). |
dif=integer (default=0) | Order of differencing of the series prior to running the test. Valid values are {0, 1, 2}. |
balance | Use balanced (across cross-sections or series) data when performing test. |
You may use one of the following keywords to specify the test:
sum (default) | Summary of all of the panel unit root tests. |
llc | Levin, Lin, and Chu. |
breit | Breitung. |
ips | Im, Pesaran, and Shin. |
adf | Fisher - ADF. |
pp | Fisher - PP. |
hadri | Hadri. |
For backward compatibility, EViews supports older forms of the exogenous specification:
const, c (default) | Include a constant in the test equation. |
trend, t | Include a constant and a linear time trend in the test equation. |
none, n | Do not include a constant or time trend (only available for the ADF and PP tests). |
For future compatibility we recommend that you use the “exog=” format.
Lag Difference Options
Specifies the number of lag difference terms to be included in the test equation. Applicable in “Summary”, LLC, Breitung, IPS, and Fisher-ADF tests. The default setting is to perform automatic lag selection using the Schwarz criteria (“lagmethod=sic”).
lagmethod=arg (default=“sic”) | Method for selecting lag lengths (number of first difference terms) to be included in the Dickey-Fuller test regressions: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “tstat” (Ng-Perron first backward significant t-statistic). |
lag=arg | Specified lag length (number of first difference terms) to be included in the regression: integer (user-specified common lag length), vector_name (user-specific individual lag length, one row per cross-section). |
maxlag=arg | Maximum lag length to consider when performing automatic lag length selection: integer (common maximum lag length), or vector_name (individual maximum lag length, one row per cross-section). The default setting produces individual maximum lags of, default= 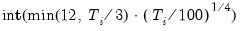 where  is the length of the cross-section. |
lagpval=arg (default=0.1) | Probability value for use in the t-statistic automatic lag selection method (when “lagmethod = tstat”). |
Kernel Options
Specifies options for computing kernel estimates of the zero-frequency spectrum (long-run covariance). Applicable to “Summary”, LLC, Fisher-PP, and Hadri tests.
hac=arg (default=“bt”) | Method of estimating the frequency zero spectrum: “bt” (Bartlett kernel), “pr” (Parzen kernel), “qs” (Quadratic Spectral kernel), |
band = arg, b=arg (default=“nw”) | Method of selecting the bandwidth: “nw” (Newey-West automatic variable bandwidth selection), “a” (Andrews automatic selection), number (user-specified common bandwidth), vector_name (user-specified individual bandwidths, one row for each cross-section). |
General options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
The command:
uroot(adf,const,lag=3,save=mout) gdp
performs an ADF test on the series GDP with the test equation including a constant term and three lagged first-difference terms. Intermediate results are stored in the matrix MOUT.
uroot(dfgls,trend,infosel=sic) ip
runs the DFGLS unit root test on the series IP with a constant and a trend. The number of lagged difference terms is selected automatically using the Schwarz criterion.
uroot(kpss,const,hac=pr,b=2.3) unemp
runs the KPSS test on the series UNEMP. The null hypothesis is that the series is stationary around a constant mean. The frequency zero spectrum is estimated using kernel methods (with a Parzen kernel), and a bandwidth of 2.3.
uroot(np,hac=ardt,infosel=maic) sp500
runs the NP test on the series SP500. The frequency zero spectrum is estimated using the OLS AR spectral estimator with the lag length automatically selected using the modified AIC.
Cross-references
See
“Unit Root Testing” for discussion of standard unit root tests performed on a single series,
“Cross-sectionally Independent Panel Unit Root Testing” for discussion of unit roots tests performed on panel structured workfiles, groups of series, or pooled data.
Specify and estimate a VAR or VEC.
Syntax
varest(options) lag_pairs endog_list [@ exog_list]
varest trend, n, options lag_pairs endog_list [@ exog_list]
The first form of the command estimates a VAR. It is the interactive command equivalent of using
ls to estimate a named VAR object (see
Var::ls for syntax and details).
The second form of the command estimates a VEC. It is the interactive command equivalent of using
ec to estimate a named VAR object (see
Var::ec for syntax and details).
Examples
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
varest 1 3 m1 gdp
estimates an unnamed unrestricted VAR with two endogenous variables (M1 and GDP), a constant and 3 lags (lags 1 through 3).
varest(noconst) 1 3 ml gdp
estimates the same VAR, but with no constant term included in the specification.
varest(method=ec) 1 4 m1 gdp tb3
estimates a VEC with four lagged first differences, three endogenous variables and one cointegrating equation using the default trend option “c”.
varest(method=ec,b,2) 1 2 4 4 tb1 tb3 tb6 @ d2 d3 d4
estimates a VEC with lagged first differences of order 1, 2, 4, three endogenous variables, three exogenous variables, and two cointegrating equations using trend option “b”.
Cross-references
Estimation using variable selection.
Syntax
varsel(options) y x1 [x2 x3 ...] @ z1 z2 z3
Specify the dependent variable followed by a list of variables to be included in the regression, but not part of the search routine, followed by an “@” symbol and a list of variables to be part of the search routine. If no included variables are required, simply follow the dependent variable with an “@” symbol and the list of search variables.
Options
method = arg | Stepwise regression method: “stepwise” (default), “uni” (uni-directional), “swap” (swapwise), “comb” (combinatorial), “gets” (auto-search/GETS), “lasso” (Lasso). |
nvars = int | Set the number of search regressors. Required for swapwise and combinatorial methods, optional for uni-directional and stepwise methods. |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Stepwise and uni-directional method options
back | Set stepwise or uni-directional method to run backward. If omitted, the method runs forward. |
tstat | Use t-statistic values as a stopping criterion. (default uses p-values). |
ftol=number (default = 0.5) | Set forward stopping criterion value. |
btol=number (default = 0.5) | Set backward stopping criterion value. |
fmaxstep=int (default = 1000) | Set the maximum number of steps forward. |
bmaxstep=int (default = 1000) | Set the maximum number of steps backward. |
tmaxstep=int (default = 2000) | Set the maximum total number of steps. |
Swapwise method options
minr2 | Use minimum R-squared increments. (Default uses maximum R-squared increments.) |
Combinatorial method options
force | Suppress the warning message issued when a large number of regressions will be performed. |
Auto-search/GETS method options
pval=number (default = 0.05) | Set the terminal condition p-value used to determine the stopping point of each search path |
nolm | Do not perform AR LM diagnostic test. |
arpval=number (default = 0.025) | Set p-value used in AR LM diagnostic test. |
arlags=int (default = 1) | Set number of lags used in AR LM diagnostic test. |
noarch | Do not perform ARCH LM diagnostic test. |
archpval=number (default = 0.025) | Set p-value used in ARCH LM diagnostic test. |
archlags=int (default = 1) | Set number of lags used in ARCH LM diagnostic test. |
nojb | Do not perform Jarque-Bera normality diagnostic test. |
jbpval=number (default = 0.025) | Set p-value used in Jarque-Bera normality diagnostic test. |
nopet | Do not perform Parsimonious Encompassing diagnostic test. |
petpval=number (default = 0.025) | Set p-value used in Parsimonious Encompassing diagnostic test. |
nogum | Do not include the general model as a candidate for model selection. |
noempty | Do not include the empty model as a candidate for model selection. |
ic =arg | Set the information criterion used in model selection: “AIC” (Akaike information criteria, default), “BIC” (Schwarz information criteria), “HQ” (Hannan-Quin criteria). |
blocks=int | Override the EViews’ determination of the number of blocks in which to split the estimation sample. |
Lasso method options
Penalty Options
ytrans=arg (default=“none”) | Scaling of the dependent variable: “none” (none), “L1” (L1), “L2” (L2), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
xtrans=arg (default=“stdpop”) | Scaling of the regressor variables: “none” (none), “L1” (L1 norm), “L2” (L2 norm), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
lambda=arg | Value of the penalty parameter. Can be a single number, list of space-delimited numbers, a workfile series object, or left blank for a EViews determined path (default). Values must be zero or greater. |
nlambdas=integer (default=100) | Number of penalty values for EViews-supplied list. |
nlambdamin=integer (default=5) | Minimum number of lambda values in the path before applying stopping rules. |
minddev=arg (default=1e-05) | Minimum change in deviance fraction to continue estimation. Truncate path estimation if relative change in deviance is smaller than this value. |
maxedev=arg (default=0.99) | Maximum of deviance explained fraction attained to terminate estimation. Truncate path estimation if fraction of null deviance explained is larger than this value. |
maxvars=arg | Maximum number of regressors in the model. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) reaches this value. |
maxvarsratio=arg | Maximum number of regressors in the model as a fraction of the number of observations. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) divided by the number of observations reaches this value. |
Cross Validation Options
cvmethod=arg (default=“kfold_cv”) | Cross-validation method: “kfold” (k-fold), “simple” (simple split), “mcarlo” (Monte Carlo), “leavepout” (leave-P-out), “leave1out” (leave-1-out), “rolling” (rolling window), “expanding” (expanding window). |
cvmeasure=arg (default=“mse”) | Cross-validation fit measure: “mse” (mean-squared error), “r2” (R‑squared), “mae” (mean absolute error), “mape” (mean absolute percentage error), “smape” (symmetric mean absolute percentage error). |
cvnfolds=arg (default=5) | Number of folds for K-fold cross-validation. For “cvmethod=kfold”. |
cvftrain=arg (default=0.8) | Proportion of data for split and Monte Carlo methods. For “cvmethod=simple” and “cvmethod=mcarlo”. |
cvnreps=arg (default=1) | Number of Monte Carlo method repetitions. For “cvmethod=mcarlo”. |
cvleaveout=arg (default=2) | Number of data points left out for leave-p-out method. For “cvmethod=leavepout”. |
cvnwindows=arg (default=4) | Number of windows for rolling window cross-validation method. For “cvmethod=rolling”. |
cvinitial=arg (default=12) | Number of initial data points in the training set for expanding cross-validation. For “cvmethod=expanding”. |
cvpregap=arg (default=0) | Number of observations between end of training set and beginning of test set. For “cvmethod=simple”, “cvmethod=rolling” and “cvmethod=expanding”. |
cvhorizon=arg (default=1) | Number of observation in the test set. For “cvmethod=rolling” and “cvmethod=expanding”. |
cvpostgap=arg (default=0) | Number of observations between end of test set and beginning of next training set for rolling window or between end of test set and end of next training set for expanding window. For “cvmethod=rolling” and “cvmethod=expanding” |
Random Number Options
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
rnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
Other Options
coefmin= vector_name, number | Vector of individual coefficient minimum values, containing negative or missing values sized to and matching the order of the variables in the specification, or a negative value for the minimum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual minimums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
coefmax= vector_name, number | Vector of individual coefficient maximum values, containing positive or missing values sized to and matching the order of the variables in the specification, or a positive value for the maximum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual maximums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
varsel(method=comb,nvars=3) y c @ x1 x2 x3 x4 x5 x6 x7 x8
performs a combinatorial search routine to search for the three variables from the set of X1, X2, ..., X8, yielding the largest R-squared in a regression of Y on a constant and those three variables.
Cross-references
Close the active or specified workfile.
Syntax
wfclose(options) [name]
wfclose allows you to close the currently active workfile. You may optionally provide the name of a workfile if you do not wish to close the active workfile. If more than one workfile is found with the same name, the one most recently opened will be closed.
Options
noerr | Do not error if a name is provided and no workfile with that name is found. |
Examples
wfclose
closes the workfile that was most recently active.
wfclose basics
closes the workfile named BASICS.
Cross-references
Compare the contents of the current workfile or page with the contents of a different workfile, page or database.
Syntax
wfcompare(options) targetspec baselinespec [@keep keeplist @drop droplist]
The command shows a list of any differences between the objects specified by targetspec and those specified by baselinespec.
targetspec should be a specification of objects in the current workfile. It should take the form of [page\]name_pattern, where the optional page\ may be used to specify a specific page of the current workfile. name_pattern is used to list the objects you wish to compare. The typical form of targetspec is simply “*”, meaning to compare all objects in the current workfile.
baselinespec should be a specification of the list of objects to compare against. It should take the form of [container::page\]name_pattern, where the optional container::page\ may be used to specify the name of the workfile or database or page containing the objects to compare against. name_pattern is used to list the objects you wish to compare against. If baselinespec is blank, the version of the current workfile stored on disk is used as the baseline.
The optional @keep and @drop lists allow you to narrow further the list of objects in targetspec by listing specific objects to compare (using @keep), or drop from the targetspec (using @drop).
Options
tol=arg | Specifies the threshold below which differences between the target and baseline values should be ignored. The threshold is specified as a fraction of the baseline value. For example, if tol=1%, one or more observations in the target object must differ from their values in the baseline object by at least one percent for the objects to be reported as different. The default tolerance is 1e-15. |
seterr | Set an error if any differences exceeding the specified tolerance were found. This option may be useful in batch programming to alert the user if a program causes unexpectedly large changes to data values. |
setsofterr | Set an error and show a list of differences if any differences exceeding the specified tolerance were found. This option may be useful in batch programming to alert the user if a program causes unexpectedly large changes to data values. |
list=key | Selects which objects should be included in the output list. key may be “a” (list objects that exist in the target but not baseline), “d” (list objects that exist in the baseline, but not target), “r” (list objects that exist in both but have different objects types), “c” (list objects that exist in both but have different frequencies), “m” (list objects that exist in both but have values that differ by more than the tolerance), “u” (list objects that exist in both and are unchanged), “s” (list objects that exist in both but cannot be compared). |
out=name | Create a table object name, containing the comparison table. |
nohead | Used with “out=” option to suppress the header row at the top of the frozen table. |
Examples
wfcompare
Compares the current workfile with the previously saved version of the workfile on disk. All pages are compared.
wfcompare *_0 *_1
compares all in the current page with names ending with “_0” with all objects whose names ends in “_1”.
wfcompare page1\* page2\*
compares all objects in page1 of the current workfile with those in page2.
wfcompare page1\* page2\* @drop GDP UNEMP
compares all objects except for the objects “GDP” and “UNEMP” in page1 of the current workfile with those in page2.
wfcompare * myfile.wf1
compares the contents of the current page with the contents of the same page in the saved workfile 'myfile.wf1'
wfcompare *\* jun2012.wf1
compares the contents of all pages in the current workfile in memory with all pages of the saved workfile “Jun2012.WF1”.
wfcompare page2\x* test.edb::y*
compares all objects in page2 of the current workfile whose name begins with the letter “x” to all objects in the database test.edb whose name begins with the letter “y”.
Cross-references
Create a new workfile. The workfile becomes the active workfile.
Syntax
wfcreate(options) frequency[(subperiod_opts)] start_date end_date [num_cross_sections]
wfcreate(options) frequency[(subperiod_opts)] start_date +num_observations
wfcreate(options) u num_observations
The first form of the command may be used to create a new regular frequency workfile with the specified frequency, start, and end date. Subperiod options may also be specified for intraweek or intraday data. See table below for a complete description for each frequency. If you include the optional
num_cross_sections, EViews will create a balanced panel page using integer identifiers for each of the cross-sections. Note that more complex panel structures may be created using
pagestruct. You may use the
@now keyword to specify the current date/time as either the
start_date or
end_date.
The second form of the command may be used to create a new regular frequency workfile with a specified frequency, start date and number of observations. The end date is calculated as the start date plus the number of periods (determined by the frequency specification) given by num_observations.
The third form of the command is used to create an unstructured workfile with the specified number of observations.
Options
wf=wf_name | Optional name for the new workfile. |
page=page_name | Optional name for the page in the new workfile. |
prompt | Force the dialog to appear from within a program. |
Arguments
The frequency argument should be specified using one of the following forms:
Sec[opt], 5Sec[opt], 15Sec[opt], 30Sec[opt] | Seconds in intervals of: 1, 5, 15, or 30 seconds, respectively. You may optionally specify days of the week and start and end times during the day using the opt parameter. See explanation of subperiod options below. |
Min[opt], 2Min[opt], 5Min[opt], 10Min[opt], 15Min[opt], 20Min[opt], 30Min[[opt] | Minutes in intervals of: 1, 2, 5, 10, 15, 20, or 30 minutes, respectively. You may optionally specify days of the week and start and end times during the day using the opt parameter. See explanation of subperiod options below. |
H[opt], 2H[opt], 4H[opt], 6H[opt], 8H[opt], 12H[[opt] | Hours in intervals of: 1, 2, 4, 6, 8, or 12 hours, respectively. You may optionally specify days of the week and start and end times during the day using the opt parameter. See explanation of subperiod options below. |
D(s, e) | Daily with arbitrary days of the week. Specify the first and last days of the week with integers s and e, where Monday is given the number 1 and Sunday is given the number 7. (Note that the “D” option used to specify a 5-day frequency prior to EViews 7.) |
D5 or 5 | Daily with five days per week, Monday through Friday. |
D7 or 7 | Daily with seven days per week. |
W | Weekly |
T | Ten-day (daily in intervals of ten). |
F | Fortnight |
BM | Bimonthly |
M | Monthly |
Q | Quarterly |
S | Semi-annual |
A or Y | Annual |
2Y, 3Y, 4Y, 5Y, 6Y, 7Y, 8Y, 9Y, 10Y, 20Y | Multi-year in intervals of: 2, 3, 4, 5, 6, 7, 8, 9, 10, or 20 years, respectively. |
Subperiod options
EViews allows for setting the days of the week and the time of day within intraday frequencies, which include seconds, minutes, and hours. For instance, you may specify hourly data between 8AM and 5PM on Monday through Wednesday. These subperiod options should follow the frequency keyword and be enclosed in parentheses.
To specify days of the week, use integers to indicate the days, where Monday is given the number 1 and Sunday is given the number 7. For example,
wfcreate(wf=storehours) 30MIN(1-6, 8:00-17:00) 1/3/2000 12/30/2000
indicates a half-hour frequency that includes Monday through Saturday from 8AM to 5PM.
To specify the start and end times, you may use either a 24 hour clock, including minutes and optionally seconds, or a 12 hour clock using AM and PM. For example, each of the following represents 8PM: 8PM, 8:00PM, 8:00:00PM, 20:00, and 20:00:00. Thus, our previous example could have been written:
wfcreate(wf=storehours) 30MIN(1-6, 8AM-5PM) 1/3/2000 12/30/2000
If you wish to include all days of the week but would like to specify a start and end time, set the date range to include all days and then specify the times. The day of the week parameter appears first and is required if you wish to supply the time of day parameters. For instance,
wfcreate(wf=storehours) 30MIN(1-7, 10AM-3PM) 1/3/2000 12/30/2000
indicates a half-hour frequency from 10AM to 3PM on all days of the week.
You may also include a time with the start and end date parameters to specify partial days at the beginning or end of the workfile. For example,
wfcreate(wf=storehours) 30MIN(1-6, 8AM-5PM) 1/3/2000 10AM 12/30/2000 2PM
creates the same workfile as above, but limits the first day, 1/3/2000, to 10AM - 5PM and the last day, 12/30/2000, to 8AM - 2PM.
Alignment options
Certain frequencies optionally allow you to specify the starting point of the frequency period. Weekly and biweekly frequencies allow you to set the day at which the week begins. Annual, semiannual and quarterly frequencies allow you to set the month at which the quarter or year begins. Setting the starting period is important if you wish to use frequency conversion to convert data from a different frequency.
To specify the start period, simply add an extra term to the frequency symbol, surrounded in parenthesis, containing the day, or month, upon which you wish the frequency to start. For example:
wfcreate w(monday) 2000 2010
creates a weekly workfile from 2000 to 2010, where each week starts on a Monday.
wfcreate a(july) 2001 2007
creates an annual workfile where each year starts in July.
Note that by default, if you do not specify a starting point, EViews will use the period of the specified start_date. To make this difference concrete, consider the commands:
wfcreate w 2000 2010
and
wfcreate w(monday) 2000 2010
Since January 1st, 2000 was a Saturday, the first command will create a weekly workfile where each week starts on a Saturday, and the first observation in the workfile will span the period January 1st-January 7th 2000.
The second command will force EViews to start weeks on a Monday, and thus the first observation will actually span the period December 27th 1999 - January 2nd 2000.
Examples
wfcreate(wf=annual, page=myproject) a 1950 2005
wfcreate(wf=unstruct, page=undated) u 1000
creates two workfiles. The first is a workfile named ANNUAL containing a single page named MYPROJECT containing annual data from 1950 to 2005; the second is a workfile named UNSTRUCT containing a single page named UNDATED with 1000 unstructured observations.
wfcreate(wf=griliches_grunfeld, page=annual) a 1935 1954 10
creates the GRILICHES_GRUNFELD workfile containing a page named “ANNUAL” with 10 cross-sections of annual data for the years 1935 to 1954.
wfcreate(wf=fourday) D(1,4) 1/3/2000 12/31/2000
specifies a daily workfile from January 3, 2000 to December 31, 2000, including only Monday through Thursday. The day range may be delimited by either a comma or a dash, such that
wfcreate(wf=fourday) D(1-4) 1/3/2000 12/31/2000
is equivalent to the previous command.
wfcreate(wf=captimes) 15SEC(2-4) 1/3/2000 12/30/2000
creates a workfile with 15 second intervals on Tuesday through Thursday only, from 1/3/2000 to 12/30/2000.
wfcreate m 1995 +30
will create a monthly workfile starting in January 1995 and ending in July 1997.
Cross-references
Change the details displayed in the current workfile window, optionally freezing the results into a table object in the current workfile.
Syntax
wfdetails(options) col1[(width1)] [col2[(width2)].....] @sort sortcol
Specify the names of the attribute columns you would like to display in the workfile details view, optionally including the width of the column in parenthesis. Widths can be specified with positive numbers, indicating a width in pixels, or negative numbers if you wish to specify the width in approximate characters.
You may use the @sort keyword at the end of the specification to indicate by which column to sort the details view.
Options
out=name | Create a table object name, containing the details view table. |
nohead | Used with “out=” option to suppress the header row at the top of the frozen table. |
Examples
wfdetails start end
shows the workfile details view, with the attribute columns “start” and “end”.
wfdetails(out=dettable) start(30) end(-10)
shows the same view, but setting the “start” column’s width to 30 pixels, and the “end” column’s width to 10 characters, and freezing the view into a table called DETTABLE.
wfdetails start end @sortcol type
sorts the details view by object type.
Cross-references
Change the workfile view to a simple object directory listing.
Syntax
wfdir
If the workfile is currently in details view, the wfdir command switches the view back to a simple directory listing.
Cross-references
Change the workfile object filter for the current workfile window.
Syntax
wffilter name_list @ object_type_list @attr(attribute_list)
Specify the names of the objects you would like to display in the workfile window using a name pattern. You may additionally specify the object type(s) using the @ delimiter and/or matching attribute value using the @attr keyword.
• The name_list is a space delimited list of names or name patterns.
• The object_type_list is a space delimited list of one or more object types or object groups. The start of the list is denoted by the '@' symbol followed by a space.
• The attribute_list is a space delimited list of attribute filters. Each applied filter is denoted by using the @attr keyword. The format of the filter has the form @attr(attr_name, attr_value).
Acceptable individual object types include:
series, group, alpha, link, sample, valmap, table, graph, text, spool, equation, var, system, logl, pool, factor, sspace, vector, sym, matrix, coef, scalar, model, user
Individual object types are also categorized into object groups:
Options
estobj | equation, var, system, logl, pool, factor, sspace |
matobj | vector, sym, matrix, coef, scalar |
viewobj | table, graph, text, spool |
dataobj | series, group, alpha, link, sample, valmap |
Examples
wffilter tab1 graph1
displays the objects tab1 and graph1 in the workfile window if they exist.
wffilter a* t*
displays in the workfile window all objects whose names begin with “a” or “t”.
wffilter * @ series
displays all the series objects in the workfile window.
wffilter g* @ series graph
displays in the workfile window the series and graph objects beginning with “g”.
wffilter * @attr("remarks","us gdp")”
displays in the workfile window the objects in the workfile whose remarks attributes are ‘us gdp”.
wffilter p* @ matobj @attr("city","") @attr("country","*US*")
displays in the workfile window the objects whose names start with “p”, are either vectors, syms, matrices, coefs, or scalars, have an empty ‘city’ attribute, and have the string ‘us’ in the ‘country’ attribute.
Cross-references
Open a workfile. Reads in a previously saved workfile from disk, or reads the contents of a foreign data source into a new workfile.
The opened workfile becomes the default workfile; existing workfiles in memory remain on the desktop but become inactive.
Syntax
wfopen [path\]source_name
wfopen(options) source_description [table_description] [variables_description]
wfopen(options) source_description [table_description] [dataset_modifiers]
where path is an optional local path or URL.
There are three basic forms of the wfopen command:
• the second form used for raw data files—Excel, Lotus, ASCII text, and binary files (
“Raw Data Formats”).
• the third form is used with the remaining source formats, which we term
dataset formats, since the data have already been arranged in named variables (
“Datasets”).
(See
“Options” for a description of the supported source formats and corresponding types.)
In all three cases, the workfile or external data source should be specified as the first argument following the command keyword and options.
• In most cases, the external data source is a file, so the source_description will be the description of the file (including local path or URL information, if necessary). Alternatively, the external data source may be the output from a web server, in which case the URL should be provided. Similarly, when reading from an ODBC query, the ODBC DSN (data source name) should be used as the source_description.
If the source_description contains spaces, it must be enclosed in (double) quotes.
For raw and dataset formats, you may use table_description to provide additional information about the data to be read:
• Where there is more than one table that could be formed from the specified external data source, a table_description may be provided to select the desired table. For example, when reading from an Excel file, an optional cell range may be provided to specify which data are to be read from the spreadsheet. When reading from an ODBC data source, a SQL query or table name must be used to specify the table of data to be read.
• In raw data formats, the table_description allows you to provide additional information regarding names and descriptions for variables to be read, missing values codes, settings for automatic format, and data range subsetting.
• When working with text or binary files, the table_description must be used to describe how to break up the file into columns and rows.
For raw and non-EViews dataset formats, you may use the dataset_modifiers specification to select the set of variables, maps (value labels), and observations to be read from the source data. The dataset_modifiers consists of the following keyword delimited lists:
[@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@selectif condition]
• The @keep and @drop keywords, followed by a list of names and patterns, are used to specify variables to be retain or dropped. Similarly, the @keepmap and @dropmap keywords followed by lists of name patterns controls the reading of value labels. The keyword @selectif, followed by an if condition (e.g., “if age>30 and gender=1”) may be used to select a subset of the observations in the original data. By default, all variables, value labels, and observations are read.
By default, all variables, maps and observations in the source file will be read.
Options
type=arg, t=arg | Optional type specification: (see table below). ODBC support is not provided in EViews Standard Edition. |
link | Link the object to the source data so that the values can be refreshed at a later time. |
wf=wf_name | Optional name for the new workfile. |
page=page_name | Optional name for the page in the new workfile. |
prompt | Force the dialog to appear from within a program. |
For the most part, you should not need to specify a “type=” option as EViews will automatically determine the type from the filename.
The following table summaries the various source formats and types, along with the corresponding “type=” keywords:
| | |
Access | dataset | “access” |
Aremos-TSD | time series database | “a”, “aremos”, “tsd” |
Binary | raw data | “binary” |
dBASE | dataset | “dbase” |
Excel (through 2003) | raw data | “excel” |
Excel 2007 (xml) | raw data | “excelxml” |
EViews Workfile | native | --- |
Gauss Dataset | dataset | “gauss” |
GiveWin/PcGive | time series database | “g”, “give” |
HTML | raw data | “html” |
Lotus 1-2-3 | raw data | “lotus” |
ODBC Dsn File | dataset | “dsn” |
ODBC Query File | dataset | “msquery” |
ODBC Data Source | dataset | “odbc” |
MicroTSP Workfile | native | “dos”, “microtsp” |
MicroTSP Mac Workfile | native | “mac” |
RATS 4.x | time series database | “r”, “rats” |
RATS Portable / TROLL | time series database | “l”, “trl” |
SAS Program | dataset | “sasprog” |
SAS Transport | dataset | “sasxport” |
SPSS | dataset | “spss” |
SPSS Portable | dataset | “spssport” |
Stata | dataset | “stata” |
Text / ASCII | raw data | “text” |
TSP Portable | time series database | “t”, “tsp” |
EViews and MicroTSP
The syntax for EViews and MicroTSP files is:
wfopen [path\]workfile_name
where path is an option local path or URL.
Examples
wfopen c:\data\macro
loads a previously saved EViews workfile “Macro.WF1” from the “data” directory in the C drive.
wfopen c:\tsp\nipa.wf
loads a MicroTSP workfile “Nipa.WF”. If you do not use the workfile type option, you should add the extension “.WF” to the workfile name when loading a DOS MicroTSP workfile. An alternative method specifies the type explicitly:
wfopen(type=dos) nipa
The command:
wfopen "<mydropboxdrive>\folder\nipa.wf1"
will open the file from the cloud location MYDROPBOXDRIVE.
Time Series Formats
The syntax for time series format files (Aremos-TSD, GiveWin/PcGive, RATS, RATS Portable/TROLL, TSP Portable) is:
wfopen(options) [path\]source_name
where path is an optional local path or URL.
If the source files contain data of multiple frequencies, the resulting workfile will be of the lowest frequency, and higher frequency data will be converted to this frequency. If you wish to obtain greater control over the workfile creation, import, or frequency conversion processes, we recommend that you open the file using
dbopen and use the database tools to create your workfile.
Aremos Example
wfopen dlcs.tsd
wfopen(type=aremos) dlcs.tsd
open the AREMOS-TSD file DLCS.
GiveWin/PcGive Example
wfopen "f:\project\pc give\data\macrodata.in7"
wfopen(type=give) "f:\project\pc give\data\macrodata"
open the PcGive file MACRODATA.
Rats Examples
wfopen macrodata.rat"
wfopen macrodata.trl
read the native RATS 4.x file MACRODATA.RAT and the RATS Portable/TROLL file “Macrodata.TRL”.
TSP Portable Example
wfopen macrodata.tsp
reads the TSP portable file “Macrodata.TSP”.
Raw Data Formats
The command for reading raw data (Excel 97-2003, Excel 2007, HTML, ASCII text, Binary, Lotus 1-2-3) is
wfopen(options) source_description [table_description] [variables_description] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@selectif condition]
where the syntax of the table_description and variables_description differs slightly depending on the type of file.
Excel and Lotus Files
The syntax for reading Excel and Lotus files is:
wfopen(options) source_description [table_description] [variables_description]
The following table_description elements may be used when reading Excel and Lotus data:
• “range = arg”, where arg is a range of cells to read from the Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the excel workbook to refer to a range or cell may be used to specify the cells to read.
• “byrow”, transpose the incoming data. This option allows you to read files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “namepos = [first|firstatt|last|lastatt|all|none|attonly|discard|custom]”, which row(s) of the column headers should be used to form the column name, and also how to use the rest. The setting “first” (or “last”) refers to the object name being in the first (or last) column header row, and all other rows as the object's description. Similarly, “firstatt” (or “lastatt”) will use the first (or last) row as the name field, but will use all others as a custom attribute. The setting “all” will concatenate all column header fields into the object's name. “none” will concatenate all column header fields into the object's description. “attonly” will save all column header fields into the object's custom attributes. “discard” will skip all header rows altogether, and “custom” will allow you to specify explicitly how to treat each column header row using the “colheadnames=” argument. The default setting is “all” if no “colheadnames=” is specified, otherwise “custom”.
• "colheadnames = ("arg1", "arg2")", required when “namepos=custom”. Specifies the name & type of each column header row. “Name” will be mapped to the object name, “Description” to the object's description field, and the rest will be stored as custom object attributes. Any blank name will cause that column header row to be skipped.
• “nonames”, the file does not contain a column header (same as “colhead=0”).
• “names=("arg1","arg2",…)”, user specified column names, where arg1, arg2, … are names of the first series, the second series, etc. when names are provided, these override any names that would otherwise be formed from the column headers.
• “descriptions=("arg1","arg2",…)”, user specified descriptions of the series. If descriptions are provided, these override any descriptions that would otherwise be read from the data.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w” (EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int| all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a “range=” argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the “na=” argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the “scan=” argument to instruct EViews to look at more rows. In addition, you may want to specify a the “na=” value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs=int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Excel Examples
wfopen "c:\data files\data.xls"
loads the active sheet of DATA.XLS into a new workfile.
wfopen(page=mypage) "c:\data files\data.xls" range="GDP data" @drop X
reads the data contained in the “GDP data” sheet of “Data.XLS” into the MYPAGE page of a new workfile. The data for the series X is dropped, and the name of the new workfile page is “GDP”.
To load the Excel file containing US Macro Quarterly data from Stock and Watson’s Introduction to Econometrics you may use the command:
wfopen http//wps.aw.com/wps/media/objects/3254/3332253/datasets2e/datasets/USMacro_Quarterly.xls
which will load the Excel file directly into EViews from the publisher’s website (as of 08/2009).
HTML Files
The syntax for reading HTML pages is:
wfopen(options) source_description [table_description] [variables_description]
The following table_description elements may be used when reading an HTML file or page:
• “table = arg”, where arg specifies which table to read in an HTML file/page containing multiple tables.
When specifying arg, you should remember that tables are named automatically following the pattern “Table01”, “Table02”, “Table03”, etc. If no table name is specified, the largest table found in the file will be chosen by default. Note that the table numbering may include trivial tables that are part of the HTML content of the file, but would not normally be considered as data tables by a person viewing the page.
• “skip = int”, where int is the number of rows to discard from the top of the HTML table.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “namepos = [first|firstatt|last|lastatt|all|none|attonly|discard|custom]”, which row(s) of the column headers should be used to form the column name, and also how to use the rest. The setting “first” (or “last”) refers to the object name being in the first (or last) column header row, and all other rows as the object's description. Similarly, “firstatt” (or “lastatt”) will use the first (or last) row as the name field, but will use all others as a custom attribute. The setting “all” will concatenate all column header fields into the object's name. “none” will concatenate all column header fields into the object's description. “attonly” will save all column header fields into the object's custom attributes. “discard” will skip all header rows altogether, and “custom” will allow you to specify explicitly how to treat each column header row using the “colheadnames=” argument. The default setting is “all” if no “colheadnames=” is specified, otherwise “custom”.
• "colheadnames = ("arg1", "arg2")", required when “namepos=custom”. Specifies the name & type of each column header row. “Name” will be mapped to the object name, “Description” to the object's description field, and the rest will be stored as custom object attributes. Any blank name will cause that column header row to be skipped.
• “nonames”, the file does not contain a column header (same as “colhead=0”).
• “names=("arg1","arg2",…)”, user specified column names, where arg1, arg2, … are names of the first series, the second series, etc. when names are provided, these override any names that would otherwise be formed from the column headers.
• “descriptions=("arg1","arg2",…)”, user specified descriptions of the series. If descriptions are provided, these override any descriptions that would otherwise be read from the data.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w”(EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a "range=" argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the "na=" argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the "scan=" argument to instruct EViews to look at more rows. In addition, you may want to specify a the "na=" value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
HTML Examples
wfopen "c:\data.html"
loads into a new workfile the data located on the HTML file “Data.HTML” located on the C:\ drive
wfopen(type=html) "http://www.tradingroom.com.au/apps/mkt/forex.ac" colhead=3, namepos=first
loads into a new workfile the data with the given URL located on the website site “http://www.tradingroom.com.au”. The column header is set to three rows, with the first row used as names for columns, and the remaining two lines used to form the descriptions.
Text and Binary Files
The syntax for reading text or binary files is:
wfopen(options) source_description [table_description] [variables_description]
If a table_description is not provided, EViews will attempt to read the file as a free-format text file. The following table_description elements may be used when reading a text or binary file:
• “ftype = [ascii|binary]” specifies whether numbers and dates in the file are stored in a human readable text (ASCII), or machine readable (Binary) form.
• “rectype = [crlf|fixed|streamed]” describes the record structure of the file:
“crlf”, each row in the output table is formed using a fixed number of lines from the file (where lines are separated by carriage return/line feed sequences). This is the default setting.
“fixed”, each row in the output table is formed using a fixed number of characters from the file (specified in “reclen= arg”). This setting is typically used for files that contain no line breaks.
“streamed”, each row in the output table is formed by reading a fixed number of fields, skipping across lines if necessary. This option is typically used for files that contain line breaks, but where the line breaks are not relevant to how rows from the data should be formed.
• “reclines =int”, number of lines to use in forming each row when “rectype=crlf” (default is 1).
• “reclen=int”, number of bytes to use in forming each row when “rectype=fixed”.
• “recfields=int”, number of fields to use in forming each row when “rectype=streamed”.
• “skip=int”, number of lines (if rectype is “crlf”) or bytes (if rectype is not “crlf”) to discard from the top of the file.
• “comment=string“, where string is a double-quoted string, specifies one or more characters to treat as a comment indicator. When a comment indicator is found, everything on the line to the right of where the comment indicator starts is ignored.
• “emptylines=[keep|drop]”, specifies whether empty lines should be ignored (“drop”), or treated as valid lines (“keep”) containing missing values. The default is to ignore empty lines.
• “tabwidth=int”, specifies the number of characters between tab stops when tabs are being replaced by spaces (default=8). Note that tabs are automatically replaced by spaces whenever they are not being treated as a field delimiter.
• “fieldtype=[delim|fixed|streamed|undivided]”, specifies the structure of fields within a record:
“Delim”, fields are separated by one or more delimiter characters
“Fixed”, each field is a fixed number of characters
“Streamed”, fields are read from left to right, with each field starting immediately after the previous field ends.
“Undivided”, read entire record as a single series.
• “quotes=[single|double|both|none]”, specifies the character used for quoting fields, where “single” is the apostrophe, “double” is the double quote character, and “both” means that either single or double quotes are allowed (default is “both”). Characters contained within quotes are never treated as delimiters.
• “singlequote“, same as “quotes = single”.
• “delim=[comma|tab|space|dblspace|white|dblwhite]”, specifies the character(s) to treat as a delimiter. “White” means that either a tab or a space is a valid delimiter. You may also use the abbreviation “d=” in place of “delim=”.
• “custom="arg1"”, specifies custom delimiter characters in the double quoted string. Use the character “t” for tab, “s” for space and “a” for any character.
• “mult=[on|off]”, to treat multiple delimiters as one. Default value is “on” if “delim” is “space”, “dblspace”, “white”, or “dblwhite”, and “off” otherwise.
• “endian = [big|little]”, selects the endianness of numeric fields contained in binary files.
• “string = [nullterm|nullpad|spacepad]”, specifies how strings are stored in binary files. If “nullterm”, strings shorter than the field width are terminated with a single zero character. If “nullpad”, strings shorter than the field width are followed by extra zero characters up to the field width. If “spacepad”, strings shorter than the field width are followed by extra space characters up to the field width.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
• “lastcol”, include implied last column. For lines that end with a delimiter, this option adds an additional column. When importing a CSV file, lines which have the delimiter as the last character (for example: “name, description, date”), EViews normally determines the line to have 3 columns. With the above option, EViews will determine the line to have 4 columns. Note this is not the same as a line containing “name, description, date”. In this case, EViews will always determine the line to have 3 columns regardless if the option is set.
A central component of the table_description element is the format statement. You may specify the data format using the following table descriptors:
• Fortran Format:
fformat=([n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)
where Type specifies the underlying data type, and may be one of the following,
I - integer
F - fixed precision
E - scientific
A - alphanumeric
X - skip
and n1, n2, ... are the number of times to read using the descriptor (default=1). More complicated Fortran compatible variations on this format are possible.
• Column Range Format:
rformat="[n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)"
where optional type is “$” for string or “#” for number, and n1, n2, n3, n4, etc. are the range of columns containing the data.
• C printf/scanf Format:
cformat="fmt"
where fmt follows standard C language (printf/scanf) format rules.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “namepos = [first|firstatt|last|lastatt|all|none|attonly|discard|custom]”, which row(s) of the column headers should be used to form the column name, and also how to use the rest. The setting “first” (or “last”) refers to the object name being in the first (or last) column header row, and all other rows as the object's description. Similarly, “firstatt” (or “lastatt”) will use the first (or last) row as the name field, but will use all others as a custom attribute. The setting “all” will concatenate all column header fields into the object's name. “none” will concatenate all column header fields into the object's description. “attonly” will save all column header fields into the object's custom attributes. “discard” will skip all header rows altogether, and “custom” will allow you to specify explicitly how to treat each column header row using the “colheadnames=” argument. The default setting is “all” if no “colheadnames=” is specified, otherwise “custom”.
• "colheadnames = ("arg1", "arg2")", required when “namepos=custom”. Specifies the name & type of each column header row. “Name” will be mapped to the object name, “Description” to the object's description field, and the rest will be stored as custom object attributes. Any blank name will cause that column header row to be skipped.
• “nonames”, the file does not contain a column header (same as “colhead=0”).
• “names=("arg1", "arg2",…)”, user specified column names, where arg1, arg2, … are names of the first series, the second series, etc. when names are provided, these override any names that would otherwise be formed from the column headers.
• “descriptions=("arg1", "arg2",…)”, user specified descriptions of the series. If descriptions are provided, these override any descriptions that would otherwise be read from the data.
• “types=("arg1","arg2",…)”, user specified data types of the series. If types are provided they will override the types automatically detected by EViews. You may use any of the following format keywords: “a” (character data), “f” (numeric data), “d” (dates), or “w” (EViews automatic detection). Note that the types appear without quotes: e.g., “types=(a,a,a)”.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file). Note: If a “range=” argument is not specified, then EViews will only scan the first five rows of data to try and determine the data format for each column. Likewise, if the “na=” argument is not specified, EViews will also try to determine possible NA values by looking for repeated values in the same rows. If the first five rows are not enough to correctly determine the data format, use the “scan=” argument to instruct EViews to look at more rows. In addition, you may want to specify a the “na=” value to override any dynamic NA value that EViews may determine on its own.
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Text and Binary File Examples (.txt, .csv, etc.)
wfopen c:\data.csv skip=5, names=(gdp, inv, cons)
reads “Data.CSV” into a new workfile page, skipping the first 5 rows and naming the series GDP, INV, and CONS.
wfopen(type=text) c:\date.txt delim=comma
loads the comma delimited data DATE.TXT into a new workfile.
wfopen(type=raw) c:\data.txt skip=8, rectype=fixed, format=(F10,X23,A4)
loads a text file with fixed length data into a new workfile, skipping the first 8 rows. The reading is done as follows: read the first 10 characters as a fixed precision number, after that, skip the next 23 characters (X23), and then read the next 4 characters as strings (A4).
wfopen(type=raw) c:\data.txt rectype=fixed, format=2(4F8,2I2)
loads the text file as a workfile using the specified explicit format. The data will be a repeat of four fixed precision numbers of length 8 and two integers of length 2. This is the same description as “format=(F8,F8,F8,F8,I2,I2,F8,F8,F8,F8,I2,I2)”.
wfopen(type=raw) c:\data.txt rectype=fixed, rformat="GDP 1-2 INV 3 CONS 6-9"
loads the text file as a workfile using column range syntax. The reading is done as follows: the first series is located at the first and second character of each row, the second series occupies the 3rd character, the third series is located at character 6 through 9. The series will named GDP, INV, and CONS.
Datasets
The syntax for reading data from the remaining sources (Access, Gauss, ODBC, SAS program, SAS transport, SPSS, SPSS portable, Stata) is:
wfopen(options) source_description table_description [@keep keep_list] [@drop drop_list] [@selectif condition]
Note that for this purpose we view Access and ODBC as datasets.
ODBC or Microsoft Access
The syntax for reading from an ODBC or Microsoft Access data source is
wfopen(options) source_description table_description [@keep keep_list] [@drop drop_list] [@selectif condition]
When reading from an ODBC or Microsoft Access data source, you must provide a table_description to indicate the table of data to be read. You may provide this information in one of two ways: by entering the name of a table in the data source, or by including an SQL query statement enclosed in double quotes.
ODBC support is not provided in EViews Standard Edition.
ODBC Examples
wfopen c:\data.dsn CustomerTable
opens in a new workfile the table named CUSTOMERTABLE from the ODBC database described in the DATA.DSN file.
wfopen(type=odbc) "my server" "select * from customers where id>30" @keep p*
opens in a new workfile with SQL query from database using the server “MY SERVER”, keeping only variables that begin with P. The query selects all variables from the table CUSTOMERS where the ID variable takes a value greater than 30.
Other Dataset Types
The syntax for reading data from the remaining sources (Gauss, SAS program, SAS transport, SPSS, SPSS portable, Stata) is:
wfopen(options) source_description [@keep keep_list] [@drop drop_list] [@selectif condition]
Note that no table_description is required.
SAS Program Example
If a data file, “Sales.DAT”, contains the following space delimited data:
AZ 110 1002
CA 200 2003
NM 90 908
OR 120 708
WA 113 1123
UT 98 987
then the following SAS program file can be read by EViews to open the data:
Data sales;
infile sales.dat';
input state $ price sales;
run;
SAS Transport Examples
wfopen(type=sasxport) c:\data.xpt
loads a SAS transport file “data.XPT” into a new workfile.
wfopen c:\inst.sas
creates a workfile by reading from external data using the SAS program statements in “Inst.SAS”. The program may contain a limited set of SAS statements which are commonly used in reading in a data file.
Stata Examples
To load a Stata file “Data.DTA” into a new workfile, dropping maps MAP1 and MAP2, you may enter:
wfopen c:\data.dta @dropmap map1 map2
To download the sports cards dataset from Stock and Watson’s Introduction to Econometrics you may use the command:
wfopen http://wps.aw.com/wps/media/objects/3254/3332253/datasets2e/datasets/Sportscards.dta
which will load the Stata dataset directly into EViews from the publisher’s website (as of 08/2009).
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
Change the workfile page order.
Syntax
wforder pagenames
where pagenames is a space delimited list of page names. If the workfile contains more pages not listed in pagenames, they will be placed at the end.
Examples
wforder annual monthly
sets the ANNUAL page as the first page in the workfile and the MONTHLY page as the second.
Refresh all links and auto-series in the active workfile. Primarily used to refresh links that use external database data.
Syntax
wfrefresh
Examples
wfrefresh
Cross-references
See also
“Series Links” for a description of link objects, and
“Auto-Updating Series”for a discussion of auto-updating series.
Save the default workfile as an EViews workfile (.wf1 file) or as a foreign file or data source.
Syntax
wfsave(options) [path\]filename
wfsave(options) source_description [nonames] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@smpl smpl_spec]
wfsave(options) source_description table_description [nonames] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@smpl smpl_spec]
saves the active workfile in the specified directory using filename. By default, the workfile is saved as an EViews workfile, but options may be used to save all or part of the active page in a foreign file or data source.
When saving to a foreign data file, the basic specification consists of a “type=” option and source_description and table_description arguments which specify the format of the foreign data file. See below for details on source_description and table_description.
The remaining optional elements specify the actual elements to be saved.
Options
Workfile (WF1) Save Options
1 | Save using single precision. |
2 | Save using double precision. |
c | Save compressed workfile (not compatible with EViews versions prior to 5.0). |
Workfile (WF2) Save Options
jf | Save JSON with formatting making it easier to read. Increases file size. |
nojf | Saves JSON without any formatting. Minimizes file size. |
gzip | Saves JSON as a compressed gzip file. Minimizes file size. |
nogzip | Saves JSON without any gzip compression (i.e., simple text file). Increases file size. |
The default workfile save settings use the global options.
Foreign Source Save Options
These options only apply when saving your file to a format other than an EViews workfile. note that some of the options only apply to specific file types.
type=arg, t=arg | Optional type specification: (see table below). ODBC support is not provided in EViews Standard Edition. |
maptype=arg | Write selected maps as: numeric (“n”), character (“c”), both numeric and character (“b”). |
nomapval | Do not write mapped values for series with attached value labels (the default is to write the mapped values if available). |
noid | Do not write observation identifiers to foreign data files (by default, EViews will include a column with the date or observation identifier). |
nonames | Do not write variable names (only application to file formats that support writing raw data without variable names). Only available in EViews 12 and later. EViews 11 and older should use nonames as an argument after the output file name. |
na=arg | String value to be used for NAs. |
attr | Include object attributes (if the output type supports it). When specified, the first column will contain attribute names and each attribute value will be displayed after the name row. |
Excel Save Options
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If the “mode=” option is not used, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
Excel 2007 Save Options
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If the “mode=” option is not used, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
cellfmt=arg | Specify whether to use EViews, pre-existing, or remove cell formatting (colors, font, number formatting when possible, column widths and row heights) for the written range. arg may be “eviews” (replace current formatting in the file with the same cell formatting in EViews), “preserve” (leave current cell formatting already in the Excel file), or “clear” (remove current formatting and do not replace). |
strlen=arg (default = 256) | Specify the maximum the number of characters written for cells containing text. Strings in cells which are longer the max, will be truncated. |
The following table summaries the various formats along with the corresponding “type=” keywords:
| | |
Access | “access” | |
Aremos-TSD | “a”, “aremos”, “tsd” | |
Binary | “binary” | |
dBASE | “dbase” | |
Excel (through 2003) | “excel” | Yes |
Excel 2007 (xml) | “excelxml” | Yes |
EViews Workfile | --- | |
Gauss Dataset | “gauss” | |
GiveWin/PcGive | “g”, “give” | |
JSON | “json” | |
JSON (legacy output generated by EViews prior to EViews 12) | “jsonlegacy” | |
EViews workfile (WF1) | “wf1” | |
EViews workfile (WF2) | “wf2” | |
Lotus 1-2-3 | “lotus” | |
ODBC Dsn File | “dsn” | |
ODBC Data Source | “odbc” | |
MicroTSP Workfile | “dos”, “microtsp” | |
MicroTSP Mac Workfile | “mac” | |
RATS 4.x | “r”, “rats” | |
RATS Portable / TROLL | “l”, “trl” | |
SAS Program | “sasprog” | |
SAS Transport | “sasxport” | |
SPSS | “spss” | |
SPSS Portable | “spssport” | |
Stata (Version 7 Format) | “stata” | |
Tableau Data Extract | tde | |
Text / ASCII | “text” | Yes |
TSP Portable | “t”, “tsp” | |
Note that if you wish to save your Excel 2007 XML file with macros enabled, you should specify the explicit filename extension “.XLSM”.
Foreign Data Descriptions
When saving to a foreign data format the base specification consists of a basic specification of source_description and table_description which specify the exact details of the format.
The command for saving as foreign data formats is
wfsave(options) source_description [table_description] [@keep keep_list] [@drop drop_list] [@keepmap keepmap_list] [@dropmap dropmap_list] [@smpl smpl_spec]
where the syntax of the table_description and variables_description differs slightly depending on the type of file.
• @keep, @drop, @keepmap, @dropmap, and @smpl arguments may be used to control what objects and observations to write.
• The nonames keyword may be used to suppress the writing of variable names in file formats that support writing raw data without variable names.
• Note that saving as a foreign data file, with the exception of JSON, will save the current workfile page only.
• The JSON type will save all series objects from all pages of the current working, ignoring any @keep, @drop, and @smpl arguments.
Excel Files
The base syntax for writing Excel files is:
wfsave(options) source_description [table_description]
where source_description is the path and name of the Excel file to be saved, and where the following table_description elements may be employed:
• “range = arg”, where arg is a range of cells to read from the Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the excel workbook to refer to a range or cell may be used to specify the cells to read.
• “byrow”, transpose the incoming data. This option allows you to read files where the series are contained in rows (one row per series) rather than columns.
Examples
Text and Binary and Other Files
The base syntax for saving other files is:
wfsave(options) source_description
where source_description is the path and name of the file to be saved.
Examples
EViews Workfile Examples
wfsave new_wf
saves the current EViews workfile as “New_wf.WF1” in the default directory.
wfsave "c:\documents and settings\my data\consump"
saves the current workfile as “Consump.WF1” in the specified path.
wfsave macro @keep gdp unemp
saves the two series GDP and UNEMP in a separate workfile, “macro.WF1” in the default directory.
wfsave macro @dropmap gdp*
saves all of the series in the current workfile, other than those that match the name pattern “gdp*” in a workfile, “macro.WF1” in the default directory.
The command:
wfsave "<mydropboxdrive>"\folder\nipa.wf1"
will save the file to the cloud location MYDROPBOXDRIVE.
Foreign Data Examples
wfsave(type=excelxml, mode=update) macro.xlsx
saves the current workfile page as a modern Excel “.XLSX” file.
wfsave(type=excelxml, mode=update) macro.xlsx range="Sheet2!a1" byrow @keep gdp unemp
will save the two series GDP and UNEMP into the existing Excel file “macro.XLSX”, specifying that the series should be written by row, starting in cell A1 on sheet Sheet2.
To save the latter file in a macro-enabled Excel 2007 file, you should specify the explicit filename extension “.XLSM”,
wfsave(type=excelxml, mode=update) macro.xlsm range="Sheet2!a1" byrow @keep gdp unemp
Alternately,
wfsave(type=excelxml, noid) macro.xlsx range="Sheet2!a1"
will save the current workfile page as the Excel file “macro.XLSX” but will not include a column of dates.
If you wish to save a column of dates in a specific date format, you can do so by first creating an alpha series in the workfile with the specified format, then saving the file with the “noid” option including that alpha series:
alpha mydates = @datestr(@date, "YYYY-MM-DD")
wfsave(type=excelxml, noid) macro.xlsm range="Sheet2!a1" @keep mydates gdp unemp
will save the series GDP and UNEMP into the Excel file “macro.XLSM” along with a date series with the format “YYYY-MM-DD”.
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
Make the selected workfile page the active workfile page.
Syntax
wfselect wfname[\pgname]
where wfname is the name of a workfile that has been loaded into memory. You may optionally provide the name of a page in the new default workfile that you wish to be made active.
Examples
wfselect myproject
wfselect myproject\page2
both change the default workfile to MYPROJECT. The first command uses the default active page, while the second changes the page to PAGE2.
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
Takes a manual snapshot of the current workfile.
Syntax
wfsnapshot(options) [label] [description]
Options
nocap | Optional flag to NOT include captured commands in snapshot meta-data. |
Both the label and description are optional arguments.
Examples
wfsnapshot “my label” “my description”
Display the workfile statistics and summary view.
Syntax
wfstats [wfname]
displays the workfile statistics and summary view showing, for each page of the specified workfile, the structure of the page, and a summary of the objects contained in the page. The specified workfile must be open. If no workfile is specified, wfstats will display results for the active workfile.
Examples
wfstats
displays the statistics view for the active workfile.
wfstats wf2
displays the statistics for the open workfile WF2.
Cross-references
See
“Workfile Summary View” for a description of the workfile statistics and summary view.
Break links in all link objects and auto-updating series (formulae) in the active workfile.
You should use some caution with this command as you will not be prompted before the links and auto-updating series are converted.
Syntax
wfunlink
Examples
wfunlink
breaks links in all pages of the active workfile
Cross-references
See
“Series Links” for a description of link objects, and
“Auto-Updating Series” for a discussion of auto-updating series.
See also
pageunlink and
unlink for page and object based unlinking, respectively.
Activates a workfile. If the workfile is currently open in EViews, then it is selected to become the default workfile. If the workfile is not currently open, it is opened.
Syntax
wfuse [path\]workfile_name[.wf1][\page_name]
The name of the workfile is specified as the argument to wfopen and may be followed by an optional page name to specify a specific page in the workfile. If the workfile is not located in the current default directory, and is not currently open in EViews, you should specify the path of the workfile along with its name. If you do specify a path, you should also include the .WF1 extension.
Examples
wfuse basics
activates the BASICS workfile. If BASICS is not currently open in EViews and is not located in the current default directory, wfuse will error.
wfuse c:\mydata\basics.wf1
activates the BASICS workfile located in “c:\mydata”.
wfuse c:\mydata\basics.wf1\page1
activates the page named PAGE1 in the BASICS workfile located in “C:\MYDATA”.
Cross-references
See
“Workfile Basics” for a discussion of workfiles.
See also
wfopen and
wfsave.
Create or change workfiles.
No longer supported; provided for backward compatibility. This command has been replaced by
wfcreate and
pageselect.
Write EViews data to a text (ASCII), Excel, or Lotus file on disk.
Creates a foreign format disk file containing EViews data. May be used to export EViews data to another program.
Unless you need to write your workfile data in transposed or in Lotus form, we recommend that you use the more powerful command for writing
a workfile page documented in
pagesave.
Syntax
write(options) [path\]filename arg1 [arg2 arg3 …]
Follow the keyword by a name for the output file and list the series to be written. The optional path name may be on the local machine, or may point to a network drive. If the path name contains spaces, enclose the entire expression in double quotation marks.
Note that EViews cannot, at present, write into an existing file. The file that you select will, if it exists, be replaced.
Options
Options are specified in parentheses after the keyword and are used to specify the format of the output file.
prompt | Force the dialog to appear from within a program. |
File type
t=dat, txt | ASCII (plain text) files. |
t=wk1, wk3 | Lotus spreadsheet files. |
t=xls | Excel spreadsheet files. |
If you omit the “t=” option, EViews will determine the type based on the file extension. Unrecognized extensions will be treated as ASCII files. For Lotus and Excel spreadsheet files specified without the “t=” option, EViews will automatically append the appropriate extension if it is not otherwise specified.
ASCII text files
na=string | Specify text string for NAs. Default is “NA”. |
names (default) / nonames | [Write / Do not write] series names. |
dates / nodates | [Write / Do not write] dates/obs. “Dates” is the default unless the “t” option for writing by series is used, in which case “nodates” is the default. |
d=arg | Specify delimiter (default is tab): “s” (space), “c” (comma). |
t | Write by series. Default is to write by obs with series in columns. |
Spreadsheet (Lotus, Excel) files
letter_number | Coordinate of the upper-left cell containing data. |
names (default) / nonames | [Write / Do not write] series names. |
dates (default) / nodates | [Write / Do not write] dates/obs. |
dates=arg | Excel format for writing date: “first” (convert to the first day of the corresponding observation if necessary), “last” (convert to the last day of the corresponding observation). |
t | Write by series. Default is to write by obs with series in columns. |
Examples
write(t=txt,na=.,d=c,dates) a:\dat1.csv hat1 hat_se1
Writes the two series HAT1 and HAT_SE1 into an ASCII file named DAT1.CSV on the A drive. The data file is listed by observations, NAs are coded as “.” (dot), each series is separated by a comma, and the date/observation numbers are written together with the series names.
write(t=txt,na=.,d=c,dates) dat1.csv hat1 hat_se1
writes the same file in the default directory.
Cross-references
See
“Exporting to a Spreadsheet or Text File” for a discussion. See
pagesave for a superior method of exporting workfile pages to foreign formats.
Close an open connection to an external application.
Syntax
xclose
xclose is used to close an open COM session with an external application.
Examples
xclose
closes the open connection.
Cross-references
See also
xopen,
xget,
xput,
xrun, and
xlog.
Retrieve data from an external application into an EViews object.
Syntax
xget(options) object @smpl sample
xget is used to retrieve data from an external COM application (support is currently provided for MATLAB and R). An existing connection to the external application must have already been opened using
xopen. The
xget command should be followed by the name of the object to retrieve, followed by an optional
@smpl keyword and a sample specification, or the name of a sample object. Including
@smpl lets you specify the observations into which the data will be retrieved.
Options
name=arg | Specify the name of the object to be created in EViews. If the name option is not specified, the created object will have the same name as the external application object that is being retrieved. |
wf=arg | Specify the workfile into which the retrieved objects will be placed. The specified workfile must be currently open inside EViews. If the wf option is not specified, the objects will be put into the current default workfile. |
page=arg | Specify the workfile page into which the retrieved objects will be placed. If the page option is not specified, the objects will be put into the current default page. |
type=arg | Specify the EViews object type to be created. arg can be “series”, “alpha”, “matrix”, “vector”, “svector”, “coef”, “sym”, “scalar” or “string”. If the type option is not set, EViews will automatically decide which object type to create. |
mode = arg (default=“merge”) | Merge options: “protect” (protect destination – only retrieve values if destination does not already exist), “merge” (prefer source -– merge only if source value is non-missing), “mergedest” (prefer destination – merge only if destination value is missing), “update” (replace all destination values in the retrieval sample with source values), “overwrite” (replace all destination values in retrieval sample with source values, and NAs outside of sample). |
R-specific options
fdata=arg | When reading a factor object, specifies how to read the factor data: as numbers (default), as labels (“labels”), as both numbers and labels (“both”). If “fdata=both”, the labels will be read into a valmap object, and the valmap will be attached to the destination series (the data target must be a series for this setting). |
fmap=arg | Name of the valmap object to hold the factor labels (when “fdata=both”). |
fmode=arg | Specifies settings for reading factor label data into a valmap: merge with preference to the existing values in the map (default), merge with preference to the factor map values (“merge”), overwrite existing valmap definitions (“overwrite”), do not alter an existing valmap (“protect”). The default method adds definitions from the factor to an existing valmap, retaining existing valmap entries that conflict with definitions from the factor. “Merge” adds definitions from the factor to an existing valmap, replacing conflicting valmap entries with definitions from the factor. “Overwrite” replaces an existing valmap specification with the factor definitions. “Protect” ensures that an existing valmap is not altered. |
MATLAB-specific options
workspace=arg (default=“base”) | MATLAB environment in which to obtain the data: “base” (base workspace), “global” (global workspace). |
Examples
xget X
retrieves an object “X” from the current open external application connection, and stores it into the current default workfile.
xget(type=vector, name=x1) Ymat
retrieves the object named “Ymat” and stores it into the current default workfile as a vector named X1.
xget(wf=mywf, type=series, name=x2) X @smpl 1990 1999
retrieves X and stores it in the MYWF workfile as a series called X2, where only the observations between 1990 and 1999 are filled in.
Cross-references
Switch on or off the external application log inside EViews.
Syntax
xlog(arg)
xlog is used to switch on or off the external COM application log window inside EViews. arg should be either “show” (to switch the log on), or “hide” (to switch it off).
Examples
xlog(hide)
switches off the log.
Cross-references
Turns off external command mode in an EViews program.
Every program line after XOFF will be treated as an EViews command.
Syntax
xoff
Examples
xopen(type=r)
xon
print(ls())
xoff
xclose
opens a connection to R, turns on external command mode, sends an R print command, turns external command mode off, and finally closes the external connection.
Cross-references
Turns on external command mode in an EViews program.
Turns on external command mode in an EViews program. Every program line after XON will be sent directly to the external application (R or MATLAB) without the need to start the command with XRUN. Call XOFF to turn this mode off.
Syntax
xoff
Examples
xopen(type=r)
xon
print(ls())
xoff
xclose
opens a connection to R, turns on external command mode, sends an R print command, turns external command mode off, and finally closes the external connection.
Cross-references
Open a connection to an external application.
Syntax
xopen(options)
xopen is used to start a COM session with an external application, either R or MATLAB. EViews can only have a single session open at a time; a session must be closed (see
xclose) before a new session can be opened.
Options
type=arg | Set the type of connection to be opened. arg may be “r” (R) or “m” (MATLAB). |
keepcurrent | If “type=” is the same as a currently open connection, keep original connection since it is already open. |
progid=arg | (optional) Set the version of MATLAB or statconnDCOM to which EViews connects when opening a session. If not specified, EViews will use the default ProgID specified in the global options. |
nolog | Do not open a session log window. |
case=arg | Specify the default case for objects in R or MATLAB using
xput. If “case=” is not provided, the default case specified in the global options will be assumed. Note that once a connection has been opened, the case option cannot be changed; you may however, use the “name=” option when using
xput to provide an explicit name. |
Note that the MATLAB ProgIDs may be of particular interest as MATLAB (R2008a and later) offers several distinct ways in which to connect to the server. The relevant ProgIDs are:
1. “MATLAB.Application”— this ProgID starts a command window version of MATLAB that was most recently used as a server (might not be the latest installed version of MATLAB).
2. “MATLAB.Application.Single”— same as (1) but starts a dedicated server so that other programs looking to use MATLAB cannot connect to your instance.
3. “MATLAB.Autoserver”—starts a command window server using the most recent version of MATLAB.
4. “MATLAB.Autoserver.Single”—same as (3) but starts a dedicated server.
5. “MATLAB.Desktop.Application”—starts the full desktop MATLAB as a server using the most recent version of MATLAB.
Each ProgID may be amended to indicate a specific version of MATLAB. For example, using the ProgID:
MATLAB.Desktop.Application.7.6
instructs EViews to use the full desktop MATLAB GUI for version R2008a (v7.6) as the Automation server.
Examples
xopen(type=m)
opens a connection to MATLAB.
xopen(type=r, case=lower)
opens a connection to R and sets the default name-case to lower.
xopen(type=m, progid=MATLAB.Desktop.Application.7.9)
opens a connection to MATLAB 7.9 running with the full desktop GUI.
Cross-references
Installs the specified R package in the current external R connection.
Syntax
xpackage(options) package_name
xpackage is only supported for external R connections (no MATLAB support) and is used to verify that a specific R package has been installed. If the package is not found, it is downloaded and installed automatically. package_name should be the name of the R package (case-sensitive).
Options
prompt | If the specified package is not found, this option forces EViews to warn the user that an R package is missing and must be installed before continuing. By default, in an EViews program, prompt is off. When running XPACKAGE from the command window, prompt is always on. |
repos=http://… | Specifies which R repository URL to use to download the package. If not specified, EViews will attempt to find the current repo site in the current R profile. If not found, and prompt is on, EViews will then ask the user to pick a repo site from the standard R CRAN mirror list. Otherwise, EViews will default to: https://cran.cnr.berkeley.edu |
Examples
xpackage rscproxy
verifies the existence of the rscproxy package. If not found, EViews will attempt to download and install it.
xopen(prompt) forecast
verifies the existence of the forecast package. If not found, EViews will first prompt the user with a timed dialog stating that the rscproxy must first be installed. Once the user clicks Yes or the dialog times out (5 seconds), EViews will then attempt to download and install it.
xopen(repos= https://muug.ca/mirror/cran) GETS
verifies the existence of the GETS package. If not found, EViews will attempt to download and install it from the specified repos link.
Cross-references
Send an EViews object to an external application.
Syntax
xput(options) ob_list drop ob1 smpl sample
xput is used to push data from EViews to an COM external application (either R or MATLAB). An existing connection to the external application must have already been opened, using xopen.
ob_list should be a space delimited list of EViews objects to push into the application. An asterisk (*) can be used as a wildcard. Objects that can be pushed include series, alphas, matrices, syms, vectors, svectors, rowvectors, coefs, scalars and strings. if the object list contains an object you do not wish to be pushed to the external application, you can use @drop followed by the name of the object.
For series and alphas, you may enter a sample to specify which observations you wish to be pushed, using @smplfollowed by the sample specification or the name of a sample object.
Options
name=arg | Specify the name or names of the objects to be created in the destination application. Multiple names may be specified using a wildcard or a space-delimited list of names. Names specified using the name option are case-sensitive so that destination objects will preserve the case of arg. If the name option is not specified, the created objects will have the same name as the EViews objects that are being pushed, with the case determined by the case established for the COM connection (see
xopen). |
mode=arg (default= “overwrite”) | Merge options: “protect” (protect destination – only put values if destination does not already exist), “overwrite” (replace all destination values with source values, and resize if necessary). |
wf=arg | Specify the workfile containing the objects to be pushed. The specified workfile must be currently open inside EViews. If the “wf=” option is not specified, the objects will be taken from the current default workfile. |
page=arg | Specify the workfile page containing the objects to be pushed. If the “page=” option is not specified, the objects will be taken from the current default page. |
map | Write value-mapped series using map labels instead of underlying values. |
R-specific options
rtype=arg | Specify the type of object to be created in R. arg can be “vector”, “ts”, “data.frame” or “list”. |
MATLAB-specific options
mtype =arg | Specify the type of object to be created in MATLAB: “matrix” (matrix object), “cell” (cell object). |
workspace=arg (default=“base”) | MATLAB environment in which to place the data: “base” (base workspace), “global” (global workspace). |
Examples
xopen(type=m)
xput(name=g) group01
opens a connection to MATLAB and then pushes the group GROUP01 to MATLAB, giving it the name G.
xopen(type=r)
xput(page=page2, rtype=vector) x @smpl 1990 1999
Opens a connection to R and pushes the series X from page PAGE2 of the current default workfile into a vector in R. Only the 10 observations for X from 1990 and 1999 are pushed into R. X will be named in R using the name “X” with the default case specified in the global options.
xopen(type=r, case=upper)
xput(rtype=data.frame, name=df1) x* @drop x2
Opens a connection to R and puts all objects whose name starts with “X”, apart from the object X2, into a data frame, named “df1”. The names of the “X*” objects will be uppercased in R.
Cross-references
See
“EViews COM Automation Client Support (MATLAB, R, Python)” for discussion. See also
“External Program Interface” for global options setting of the default case for names.
Run a command in an external application.
Syntax
xrun command
xrun is used to run a command in an external COM application (either R or MATLAB). An existing connection to the external application must have already been opened using xopen. xrun should be followed by the command you wish to run.
Examples
xopen(type=m, case=upper)
xput Y
xput XS
xrun beta = inv(XS'*XS)*XS'*Y
opens a connection to MATLAB, sends the series Y and the group XS to MATLAB, then runs a command which will compute the least squares coefficients from a regression of Y on XS.
The commands
xopen(type=r, case=upper)
xput(rtype=data.frame, name=cancer) age drug2 drug3 studytim
xrun z<-glm(STUDYTIM~AGE+1+DRUG2+DRUG3, family=Gamma(link=log),data=cancer)
send data to R and estimate GLM model.
Note that the statconnDCOM package does not always automatically capture all of your R output. Consequently, you may find that using xrun to issue a command that displays output in R may only return a subset of the usual output to your log window. In the most extreme case, you may see a simple “OK” message displayed in your log window. To instruct statconnDCOM to show all of the output, you should use enclose your command inside an explicit print statement in R. Thus, to display the contents of a matrix X, you must issue the command
xrun print(X)
instead of the more natural
xrun X
Cross-references
 or
or 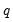 is 9; values above will be set to 9. The minimum value for
is 9; values above will be set to 9. The minimum value for 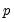 is 1. The minimum value for
is 1. The minimum value for 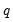 is 0. If either
is 0. If either  or
or 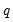 is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default.
is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default. 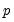 or
or 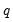 is 9; values above will be set to 9. The minimum value for
is 9; values above will be set to 9. The minimum value for 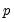 is 1. The minimum value for
is 1. The minimum value for 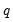 is 0. If either
is 0. If either  or
or 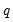 is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default.
is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default. 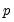 and
and 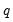 options should not be specified, and if provided, will be ignored.
options should not be specified, and if provided, will be ignored. .
.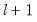 versus
versus 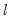 breaks), “seqall” (sequential test of all possible
breaks), “seqall” (sequential test of all possible 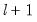 versus
versus  breaks), “glob” (tests of global
breaks), “glob” (tests of global 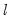 vs. no breaks), “globplus1” (tests of
vs. no breaks), “globplus1” (tests of 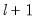 versus
versus 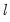 globally determined breaks), “globinfo” (information criteria evaluation), “user” (user-specified break dates).
globally determined breaks), “globinfo” (information criteria evaluation), “user” (user-specified break dates).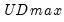 (“udmax”),
(“udmax”), 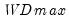 (“wdmax”).
(“wdmax”).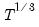 .
.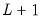 versus
versus 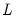 tests to determine the optimal breaks in a model regressing M1 on the breaking variables C and UNEMP.
tests to determine the optimal breaks in a model regressing M1 on the breaking variables C and UNEMP.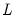 versus none tests to determine the breaks. The selected break will be the highest significant number of breaks.
versus none tests to determine the breaks. The selected break will be the highest significant number of breaks.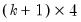 matrix, where
matrix, where 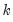 is the number of endogenous variables in the VAR. The first column contains the eigenvalues, the second column contains the maximum eigenvalue statistics, the third column contains the trace statistics, and the fourth column contains the log likelihood values. The
is the number of endogenous variables in the VAR. The first column contains the eigenvalues, the second column contains the maximum eigenvalue statistics, the third column contains the trace statistics, and the fourth column contains the log likelihood values. The 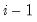 . The last row is filled with NAs, except the last column which contains the log likelihood value of the unrestricted (full rank) model.
. The last row is filled with NAs, except the last column which contains the log likelihood value of the unrestricted (full rank) model. 
 is the number of coefficients in the cointegrating equation. Applicable when “lag=a”.
is the number of coefficients in the cointegrating equation. Applicable when “lag=a”.
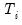 is the length of the cross-section.
is the length of the cross-section. Pearson correlation matrix for the three series HEIGHT, WEIGHT, and AGE.
Pearson correlation matrix for the three series HEIGHT, WEIGHT, and AGE. Pearson covariance matrix for the three series HEIGHT, WEIGHT, and AGE.
Pearson covariance matrix for the three series HEIGHT, WEIGHT, and AGE. , or to place inequality restrictions on the coefficient values, you may do so using special expressions of the form:
, or to place inequality restrictions on the coefficient values, you may do so using special expressions of the form: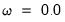 .
.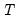 and the number of potential regressors
and the number of potential regressors 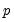 .
.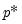 where
where 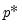 the number of non-zero
the number of non-zero  .
. 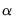 equal to the default of 0.9, no regressor or dependent variable scaling, automatically determined 100-element lambda path with minimum lambda of 0.0001 times the maximum value, using the default K-fold cross-validation with 5 folds with an MSE objective and a random generator seed of 513255899 to determine the optimal value.
equal to the default of 0.9, no regressor or dependent variable scaling, automatically determined 100-element lambda path with minimum lambda of 0.0001 times the maximum value, using the default K-fold cross-validation with 5 folds with an MSE objective and a random generator seed of 513255899 to determine the optimal value.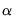 equal to the 0.75 using rolling window cross-validation and SMAPE cross-validation.
equal to the 0.75 using rolling window cross-validation and SMAPE cross-validation.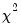 statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”).
statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”).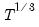 .
.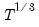 .
.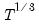 .
.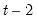 to 1.
to 1.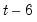 to
to 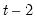 , and ordinary instruments X(-1) and Y(-1).
, and ordinary instruments X(-1) and Y(-1). 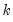 in the K‑class estimator. If omitted, LIML is performed, and
in the K‑class estimator. If omitted, LIML is performed, and 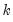
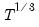 .
. matrix.
matrix. 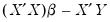 , and asking the optimizer to find the values of
, and asking the optimizer to find the values of  that minimize the square of those residuals:
that minimize the square of those residuals: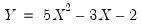
 , the covariance matrix of distribution.
, the covariance matrix of distribution. . This form is more efficient when performing multiple draws from the same distribution (compute the Cholesky once, but sample many times).
. This form is more efficient when performing multiple draws from the same distribution (compute the Cholesky once, but sample many times). . This form is more efficient than explicitly inverting
. This form is more efficient than explicitly inverting 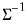 to supply
to supply 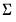 .
.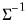 . This form combines the efficiencies of the above forms.
. This form combines the efficiencies of the above forms.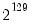
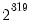
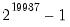
 draws)
draws) element of the matrix corresponds to
element of the matrix corresponds to 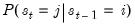 .
. and LR test statistics, as well as the test regression.
and LR test statistics, as well as the test regression. versus
versus  thresholds), “seqall” (sequential test of all possible
thresholds), “seqall” (sequential test of all possible  versus
versus 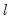 thresholds), “glob” (tests of global
thresholds), “glob” (tests of global 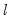 vs. no thresholds), “globplus1” (tests of
vs. no thresholds), “globplus1” (tests of  versus
versus  globally determined thresholds), “globinfo” (information criteria evaluation)., “fixedseq” (fixed number of sequentially determined thresholds), “fixedglob” (fixed number of globally determined thresholds), “user” (user-specified thresholds)
globally determined thresholds), “globinfo” (information criteria evaluation)., “fixedseq” (fixed number of sequentially determined thresholds), “fixedglob” (fixed number of globally determined thresholds), “user” (user-specified thresholds)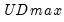 ("udmax"),
("udmax"), 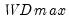 ("wdmax").
("wdmax").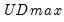 ("udmax"),
("udmax"), 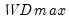 ("wdmax").
("wdmax"). .
.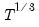 .
.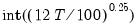
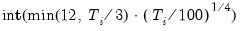
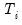 is the length of the cross-section.
is the length of the cross-section.