Matrix
Matrix (two-dimensional array).
Matrix Declaration
There are several ways to create a matrix object. You can enter the matrix keyword (with an optional row and column dimension) followed by a name:
matrix scalarmat
matrix(10,3) results
Alternatively, you can combine a declaration with an assignment statement, in which case the new matrix will be sized accordingly.
Lastly, a number of object procedures create matrices.
Matrix Views
cor correlation matrix by columns.
cov covariance matrix by columns.
display display table, graph, or spool in object window.
freq 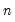
-way contingency table.
label label information for the matrix.
pcomp principal components analysis of the columns in a matrix.
sheet spreadsheet view of the matrix.
stats descriptive statistics for each column of the matrix.
testbtw tests of equality for mean, median, or variance between series in group.
Matrix Procs
clearhist clear the contents of the history attribute.
copy creates a copy of the matrix.
distdata save a matrix containing distribution plot data computed from the matrix.
export save matrix as Excel 2007 XLSX, CSV, tab-delimited ASCII text, RTF, HTML, Enhanced Metafile, PDF, TEX, or MD file on disk.
fill fill the elements of the matrix.
import imports data from a foreign file into the matrix object.
label set label information for the matrix.
makepcomp save the scores from a principal components analysis of the matrix.
olepush push updates to OLE linked objects in open applications.
read (deprecated) import data from disk.
resample resample from rows of the matrix.
resize resize the matrix object.
setattr set the value of an object attribute.
setformat set the display format for the matrix spreadsheet.
setindent set the indentation for the matrix spreadsheet.
setjust set the horizontal justification for all cells in the spreadsheet view of the matrix object.
setwidth set the column width in the matrix spreadsheet.
showlabels displays the custom row and column labels of a matrix spreadsheet.
write export data to disk.
Matrix Graph Views
Graph creation views are discussed in detail in
“Graph Creation Command Summary”.
area area graph of the columns in the matrix.
bar bar graph of each column.
hilo high-low(-open-close) chart.
line line graph of each column.
qqplot quantile-quantile graph.
scat scatter diagrams of the columns of the matrix.
scatmat matrix of all pairwise scatter plots.
Matrix Data Members
String values
@attr("arg") string containing the value of the arg attribute, where the argument is specified as a quoted string.
@collabels string containing the column labels of the matrix.
@description string containing the Matrix object’s description (if available).
@detailedtype string with the object type: “MATRIX”.
@displayname string containing the Matrix object’s display name. If the Matrix has no display name set, the name is returned.
@name string containing the Matrix object’s name.
@remarks string containing the Matrix object’s remarks (if available).
@rowlabels string containing the row labels of the matrix.
@type string with the object type: “MATRIX”.
@updatetime string representation of the time and date at which the Matrix was last updated.
Scalar values
(i,j) (i,j)-th element of the matrix. Simply append “(i, j)” to the matrix name (without a “.”).
@cols number of columns in the matrix.
@rows number of rows in the matrix.
Matrix values
@col(arg) Returns the columns defined by arg. arg may be an integer, vector of integers, string, or svector of strings. Integers correspond to column numbers so that, for example, arg = 2 specifies the second column. Strings correspond to column labels so that arg = "2" specifies the first column labeled “2”.
@diag vector containing the diagonal elements of the matrix.
@dropboth(arg1, arg2) Returns the matrix with the rows defined by arg1 and columns defined by arg2 removed. The args may be integers, vectors of integers, strings, or svectors of strings. Integers correspond to row or column numbers so that, for example, arg1 = 2 specifies the second row. Strings correspond to row or column labels so that arg2 = "2" specifies the first column labeled “2”.
@dropcol(arg) Returns the matrix with the columns defined by arg removed. arg may be an integer, vector of integers, string, or svector of strings. Integers correspond to column numbers so that, for example, arg = 2 specifies the second column. Strings correspond to column labels so that arg = "2" specifies the first column labeled “2”.
@droprow(arg) Returns the matrix with rows defined by arg removed. arg may be an integer, vector of integers, string, or svector of strings. Integers correspond to row numbers so that, for example, arg = 2 specifies the second row. Strings correspond to row labels so that arg = "2" specifies the first row labeled “2”.
@icol(arg) Returns the indices for the columns defined by arg where arg is a string or svector of strings. The strings correspond to column labels so that arg = "2" specifies the first column labeled “2”.
@irow(arg) Returns the indices for the rows defined by arg where arg is a string or svector of strings. The strings correspond to row labels so that arg = "2" specifies the first row labeled “2”.
@row(arg) Returns the rows defined by arg. arg may be an integer, vector of integers, string, or svector of strings. Integers correspond to row numbers so that, for example, arg = 2 specifies the second row. Strings correspond to row labels so that arg = "2" specifies the first row labeled “2”.
@sub(arg1, arg2) Returns the matrix with rows defined by arg1 and columns with defined by arg2. The args may be integers, vectors of integers, strings, or svectors of strings. Integers correspond to row or column numbers so that, for example, arg1 = 2 specifies the second row. Strings correspond to row or column labels so that arg2 = "2" specifies the first column labeled “2”.
@t transpose of the matrix.
Matrix Examples
The following assignment statements create and initialize matrix objects,
matrix copymat=results
matrix covmat1=eq1.@coefcov
matrix(5,2) count
count.fill 1,2,3,4,5,6,7,8,9,10
as does the equation procedure:
eq1.makecoefcov covmat2
You can declare and initialize a matrix in one command:
matrix(10,30) results=3
matrix(5,5) other=results1
Graphs and covariances may be generated for the columns of the matrix,
copymat.line
copymat.cov
and statistics computed for the rows of a matrix:
matrix rowmat=@transpose(copymat)
rowmat.stats
You can use explicit indices to refer to matrix elements:
scalar diagsum=cov1(1,1)+cov1(2,2)+cov(3,3)
Clear the column labels in a matrix object.
Syntax
matrix_name.clearcollabels
Examples
mat1.clearcollabels
clears the custom column labels from the matrix MAT1.
Cross-references
Clear the contents of the history attribute for matrix objects.
Removes the matrix’s history attribute, as shown in the label view of the matrix.
Syntax
matrix_name.clearhist
Examples
m1.clearhist
m1.label
The first line removes the history from the matrix M1, and the second line displays the label view of M1, including the now blank history field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Clear the contents of the remarks attribute.
Removes the matrix’s remarks attribute, as shown in the label view of the matrix.
Syntax
matrix_name.clearremarks
Examples
m1.clearremarks
m1.label
The first line removes the remarks from the matrix M1, and the second line displays the label view of M1, including the now blank remarks field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Clear the row labels in a matrix object.
Syntax
matrix_name.clearrowlabels
Examples
mat1.clearrowlabels
clears the custom row labels from the matrix MAT1.
Cross-references
Creates a copy of the matrix.
Creates either a named or unnamed copy of the matrix.
Syntax
matrix_name.copy
matrix_name.copy dest_name
Examples
m1.copy
creates an unnamed copy of the matrix M1.
m1.copy m2
creates M2, a copy of the matrix M1.
Cross-references
Compute covariances, correlations, and other measures of association for the columns in a matrix.
You may compute measures related to Pearson product-moment (ordinary) covariances and correlations, Spearman rank covariances, or Kendall’s tau along with test statistics for evaluating whether the correlations are equal to zero.
Syntax
matrix_name.cor(options) [keywords [@partial z1 z2 z3...]]
You should specify keywords indicating the statistics you wish to display from the list below, optionally followed by the keyword @partial and the name of a conditioning matrix. The columns should contain the conditioning variables, and the number of rows should match the original matrix.
You may specify keywords from one of the four sets (Pearson correlation, Spearman correlation, Kendall’s tau, Uncentered Pearson) corresponding the computational method you wish to employ. (You may not select keywords from more than one set.)
If you do not specify
keywords, EViews will assume “corr” and compute the Pearson correlation matrix. Note that
Matrix::cor is equivalent to the
Matrix::cov command with a different default setting.
Pearson Correlation
cov | Product moment covariance. |
corr | Product moment correlation. |
sscp | Sums-of-squared cross-products. |
stat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
prob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Spearman Rank Correlation
rcov | Spearman’s rank covariance. |
rcorr | Spearman’s rank correlation. |
rsscp | Sums-of-squared cross-products. |
rstat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
rprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Kendall’s tau
taub | Kendall’s tau-b. |
taua | Kendall’s tau-a. |
taucd | Kendall’s concordances and discordances. |
taustat | Kendall’s score statistic for evaluating whether the Kendall’s tau-b measure is zero. |
tauprob | Probability under the null for the score statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Uncentered Pearson
ucov | Product moment covariance. |
ucorr | Product moment correlation. |
usscp | Sums-of-squared cross-products. |
ustat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
uprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Note that cases, obs, and wgts are available for each of the methods.
Options
wgt=name (optional) | Name of vector containing weights. The number of rows of the weight vector should match the number of rows in the original matrix. |
wgtmethod=arg (default = “sstdev” | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
multi=arg (default=“none”) | Adjustment to p-values for multiple comparisons: none (“none”), Bonferroni (“bonferroni”), Dunn-Sidak (“dunn”). |
outfmt=arg (default=“single”) | Output format: single table (“single”), multiple table (“mult”), list (“list”), spreadsheet (“sheet”). Note that “outfmt=sheet” is only applicable if you specify a single statistic keyword. |
out=name | Basename for saving output. All results will be saved in Sym matrices named using keys (“COV”, “CORR”, “SSCP”, “TAUA”, “TAUB”, “CONC” (Kendall’s concurrences), “DISC” (Kendall’s discordances), “CASES”, “OBS”, “WGTS”) appended to the basename (e.g., the covariance specified by “out=my” is saved in the Sym matrix “MYCOV”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
mat1.cor
displays a
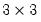
Pearson correlation matrix for the columns series in
MAT1.
mat1.cor corr stat prob
displays a table containing the Pearson correlation, t-statistic for testing for zero correlation, and associated p-value, for the columns in MAT1.
mat1.cor(pairwise) taub taustat tauprob
computes the Kendall’s tau-b, score statistic, and p-value for the score statistic, using samples with pairwise missing value exclusion.
grp1.cor(out=aa) cov
computes the Pearson covariance for the columns in MAT1 and saves the results in the symmetric matrix object AACO.
Cross-references
See also
Matrix::cov. For simple forms of the calculation, see
@cor, and
@cov.
Compute covariances, correlations, and other measures of association for the columns in a matrix.
You may compute measures related to Pearson product-moment (ordinary) covariances and correlations, Spearman rank covariances, or Kendall’s tau along with test statistics for evaluating whether the correlations are equal to zero.
Syntax
matrix_name.cov(options) [keywords [@partial z1 z2 z3...]]
You should specify keywords indicating the statistics you wish to display from the list below, optionally followed by the keyword @partial and the name of a conditioning matrix. The columns should contain the conditioning variables, and the number of rows should match the original matrix.
You may specify keywords from one of the four sets (Pearson correlation, Spearman correlation, Kendall’s tau, Uncentered Pearson) corresponding the computational method you wish to employ. (You may not select keywords from more than one set.)
If you do not specify
keywords, EViews will assume “cov” and compute the Pearson covariance matrix. Note that
Matrix::cov is equivalent to the
Matrix::cor command with a different default setting.
Pearson Correlation
cov | Product moment covariance. |
corr | Product moment correlation. |
sscp | Sums-of-squared cross-products. |
stat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
prob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Spearman Rank Correlation
rcov | Spearman’s rank covariance. |
rcorr | Spearman’s rank correlation. |
rsscp | Sums-of-squared cross-products. |
rstat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
rprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Kendall’s tau
taub | Kendall’s tau-b. |
taua | Kendall’s tau-a. |
taucd | Kendall’s concordances and discordances. |
taustat | Kendall’s score statistic for evaluating whether the Kendall’s tau-b measure is zero. |
tauprob | Probability under the null for the score statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Uncentered Pearson
ucov | Product moment covariance. |
ucorr | Product moment correlation. |
usscp | Sums-of-squared cross-products. |
ustat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
uprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Note that cases, obs, and wgts are available for each of the methods.
Options
wgt=name (optional) | Name of vector containing weights. The number of rows of the weight vector should match the number of rows in the original matrix. |
wgtmethod=arg (default = “sstdev”) | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction for the mean (for centered specifications), and any partial conditioning variables. |
multi=arg (default=“none”) | Adjustment to p-values for multiple comparisons: none (“none”), Bonferroni (“bonferroni”), Dunn-Sidak (“dunn”). |
outfmt=arg (default= “single”) | Output format: single table (“single”), multiple table (“mult”), list (“list”), spreadsheet (“sheet”). Note that “outfmt=sheet” is only applicable if you specify a single statistic keyword. |
out=name | Basename for saving output. All results will be saved in Sym matrices named using keys (“COV”, “CORR”, “SSCP”, “TAUA”, “TAUB”, “CONC” (Kendall’s concurrences), “DISC” (Kendall’s discordances), “CASES”, “OBS”, “WGTS”) appended to the basename (e.g., the covariance specified by “out=my” is saved in the Sym matrix “MYCOV”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
mat1.cov
displays a
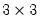
Pearson covariance matrix for the columns series in
MAT1.
mat1.cov corr stat prob
displays a table containing the Pearson covariance, t-statistic for testing for zero correlation, and associated p-value, for the columns in MAT1.
mat1.cov(pairwise) taub taustat tauprob
computes the Kendall’s tau-b, score statistic, and p-value for the score statistic, using samples with pairwise missing value exclusion.
mat1.cov(out=aa) cov
computes the Pearson covariance for the columns in MAT1 and saves the results in the symmetric matrix object AACO.
Cross-references
See also
Matrix::cor. For simple forms of the calculation, see
@cor, and
@cov.Display table, graph, or spool output in the matrix object window.
Display the contents of a table, graph, or spool in the window of the matrix object.
Syntax
matrix_name.display object_name
Examples
matrix1.display tab1
Display the contents of the table TAB1 in the window of the object MATRIX1.
Cross-references
Most often used in constructing an EViews Add-in. See
“Custom Object Output”.
Display names for matrix objects.
Attaches a display name to a matrix object which may be used to label output in place of the standard matrix object name.
Syntax
matrix_name.displayname display_name
Display names are case-sensitive, and may contain a variety of characters, such as spaces, that are not allowed in matrix object names.
Examples
m1.displayname Hours Worked
m1.label
The first line attaches a display name “Hours Worked” to the matrix object M1, and the second line displays the label view of M1, including its display name.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Save a matrix containing distribution plot data computed from the matrix.
Saves the data used to display the kernel regression, nearest neighbor regression, or empirical quantile-quantile plot to the workfile.
Syntax
matrix_name.distdata(dtype=dist_type, dist_options) matrix_name_pattern
saves the distribution plot data specified by dist_type where dist_type must be one of the following keywords:
kernfit | Kernel regression (default). |
nnfit | Nearest neighbor (local) regression. |
empqq | Empirical quantile-quantile plot. |
The matrix_name_pattern is used to define a naming pattern for the output matrices; if the pattern is “NAME”, the resulting matrices will be named “NAME01”, “NAME02”, … and so on, using the next available name.
Options
For the first two types (“kernfit” and “nnfit”),
dist_options are any of the distribution type-specific options described in
“Kernfit Options” and
“Nnfit Options”, respectively. The empirical quantile-quantile plot type (“empqq”) takes the options described in
qqplot under
“Empirical Options”.
Note that the graph display specific options such as “fill,” “nofill,” “leg,” and “noline” are not relevant for this procedure.
In addition, you may use the “mult” option to specify multiple series handling
mult = mat_type | Multiple series or column handling: where mat_type may be: “pairs” or “p” - pairs, “mat” or “m” - scatterplot matrix, “lower” or “l” - lower triangular matrix. |
and the “prompt” option to force the dialog display
prompt | Force the dialog to appear from within a program. |
Examples
If MAT1 is a matrix with four columns,
mat1.distdata(mult=first, dtype=kernel, k=e, ngrid=100) m
then creates three matrices, M01, M02 and M03, where the M01 contains the kernel fit (with an Epanechnikov kernel and 100 grid points) of the second column of MAT1 on the first column, M02 contains the fit of the first column on the third column, and M03 contains the kernel fit of column 1 on column 4.
mat1.distdata(mult=pairs, dtype=local, b=0.3, d=1, neval=100, s) n
creates two matrices, N1 and N2, where N1 contains the nearest neighbor fit of column 1 on column 2 computed using a bandwidth of 0.3 and polynomial degree of 1, 100 evaluation points and symmetric neighbors, and N2 contains the data for the nearest neighbor fit of column 3 on column 4.
If we extract a new matrix MAT2 containing first three columns of MAT1, the commands
matrix mat2 = mat1.@col(@range(1, 3))
mat2.distdata(mult=all, dtype=empqq, q=r) mat
creates 3 matrices; MAT01, MAT02, and MAT03, where MAT01 contains the empirical quantile-quantile for columns 1 and 2, computed using the rankit quantile method, MAT02 contains the qq-plot data for columns 1 and 3, and MAT03 contains the qq-plot data for columns 2 and 3.
Cross-references
For a description of distribution graphs and quantile-quantile graphs, see
“Auxiliary Graph Types”.
Export matrix to disk as an Excel 2007 XLSX, CSV, tab-delimited ASCII text, RTF, HTML, Enhanced Metafile, LaTeX, PDF, or Markdown file.
Syntax
matrix_name.export(options) [path\]file_name
Follow the keyword with a name for the file. file_name may include the file type extension, or the file type may be specified using the “t=” option.
If an explicit path is not specified, the file will be stored in the default directory, as set in the global options.
The base syntax for writing Excel 2007 files is:
matrix_name.export(options) [path\]file_name [table_description]
where the table_description may contain:
• “range = arg”, where arg is top left cell of the destination Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the Excel workbook to refer to a range or cell may be used to specify the cells to read.
Options
t=file_type (default=“csv”) | Specifies the file type, where file_type may be one of: “excelxml” (Excel 2007 (xml)),“csv” (CSV - comma-separated), “rtf” (Rich-text format), “txt” (tab-delimited text), “html” (HTML - Hypertext Markup Language), “emf” (Enhanced Metafile), “pdf” (PDF - Portable Document Format), “tex” (LaTeX), or “md” (Markdown). Files will be saved with the “.xlsx”, “.csv”, “.rtf”, “.txt”, “.htm”, “.emf”, “.pdf”, “.tex”, or “.md” extensions, respectively. |
s=arg | Scale size, where arg is from 5 to 200, representing the percentage of the original table size (only valid for HTML or RTF files). |
n=string | Replace all cells that contain NA values with the specified string. “NA” is the default. |
h / -h | Include(/do not include) column and row headers. The default is to not include the headers |
prompt | Force the dialog to appear from within a program. |
PDF Options
landscape | Save in landscape mode (the default is to save in portrait mode). |
size=arg (default=“letter”) | Page size: “letter”, “legal”, “a4”, and “custom”. |
width=number (default=8.5) | Page width in inches if “size=custom”. |
height=number (default=11) | Page height in inches if “size=custom”. |
leftmargin=number (default=0.5) | Left margin width in inches. |
rightmargin=number (default = 0.5) | Right margin width in inches. |
topmargin=number (default=1) | Top margin width in inches. |
bottommargin= number (default = 1) | Bottom margin width in inches. |
LaTeX Options
texspec / -texspec | [Include / Do not include] the full LaTeX documentation specification in the LaTeX output. The default behavior is taken from the global default settings. |
Excel Options
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If the “mode=” option is not used, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
Excel 2007 Options
mode=arg | Specify whether to create a new file, overwrite an existing file, or update an existing file. arg may be “create” (create new file only; error on attempt to overwrite) or “update” (update an existing file, only overwriting the area specified by the range= table_description). If the “mode=” option is not used, EViews will create a new file, unless the file already exists in which case it will overwrite it. Note that the “mode=update” option is only available for Excel in 1) Excel versions through 2003, if Excel is installed, and 2) Excel 2007 (xml). Note: Excel does not need to be installed for Excel 2007 writing. |
cellfmt=arg | Specify whether to use EViews, pre-existing, or remove cell formatting (colors, font, number formatting when possible, column widths and row heights) for the written range. arg may be “eviews” (replace current formatting in the file with the same cell formatting in EViews), “preserve” (leave current cell formatting already in the Excel file), or “clear” (remove current formatting and do not replace). |
strlen=arg (default = 256) | Specify the maximum the number of characters written for cells containing text. Strings in cells which are longer the max, will be truncated. |
Examples
The command:
matrix1.export mymatrix
exports the data in MATRIX1 to a CSV file named “mymatrix.CSV” in the default directory.
matrix1.export(h, t=csv, n="NaN") mymatrix
saves the contents of MATRIX1 along with the column and row headers to a CSV (comma separated value) file named “mymatrix.CSV” and writes all NA values as “NaN”.
matrix1.export(h, t=html, s=50) mymatrix
exports the data in MATRIX1 along with the column and row headers to a HTML file named “mymatrix.HTM” at half of the original size.
matrix1.export(n=".", r=B) mymatrix
saves the data in the second column to a CSV file named “mymatrix.CSV”, and writes all NA values as “.”.
matrix1.export(t=excelxml, cellfmt=clear, mode=update) mymatrix range=Country!b5
writes the data in MATRIX1 to the preexisting “mymatrix.XLSX” Excel file to the “Country” sheet at cell B5, where all cell formatting is cleared.
Cross-references
Fill a matrix object with specified values.
Syntax
matrix_name.fill(options) n1[, n2, n3 …]
Follow the keyword with a list of values to place in the matrix object. Each value should be separated by a comma.
Running out of values before the object is completely filled is not an error; the remaining cells or observations will be unaffected, unless the “l” option is specified. If, however, you list more values than the object can hold, EViews will not modify any observations and will return an error message.
Options
l | Loop repeatedly over the list of values as many times as it takes to fill the object. |
o=integer (default=1) | Fill the object starting from the specified element. Default is the first element. |
b=arg (default=“c”) | Matrix fill order: “c” (fill the matrix by column), “r” (fill the matrix by row). |
Examples
The commands,
matrix(2,2) m1
matrix(2,2) m2
m1.fill 1, 0, 1, 2
m2.fill(b=r) 1, 0, 1, 2
create the matrices:
 | (1.2) |
Cross-references
See
“Matrix Language” for a detailed discussion of vector and matrix manipulation in EViews.
Compute frequency tables for columns of a matrix.
The freq command performs a one-way or N-way frequency tabulation.
• When used with a matrix containing a single column, freq performs a one-way frequency tabulation.
• When used with a matrix containing multiple columns, freq produces an N-way frequency tabulation for all of the columns in the matrix.
Frequencies are computed for all of the rows of the matrix. Rows with NAs are dropped unless included by option. You may use options to control automatic binning (grouping) of values and the order of the entries of the table.
Syntax
matrix_name.freq(options)
Options
Options common to both one-way and N-way frequency tables
dropna (default) / keepna | [Drop/Keep] NA as a category. |
v=integer (default=1000) | Make bins if the number of distinct values or categories exceeds the specified number. |
nov | Do not make bins on the basis of number of distinct values; ignored if you set “v=integer.” |
a=number | (optional) Make bins if average count per distinct value is less than the specified number. |
b=integer (default=50) | Maximum number of categories to bin into if performing automatic binning. |
n, obs, count (default) | Display frequency counts. |
nocount | Do not display frequency counts. |
nolimit | Remove prompt warning for continuing when the total number of cells is very large. |
sort=arg (default=“lohi”) | Sort order for entries in the frequency table: high data value to low ("hilo"), low data value to high ("lohi" –default), high frequency to low ("freqhilo"), low frequency to high ("freqlohi"). |
prompt | Force the dialog to appear from within a program. |
p | Print the table. |
Options for one-way tables
total (default) / nototal | [Display / Do not display] totals. |
pct (default) / nopct | [Display / Do not display] percent frequencies. |
cum (default) / nocum | (Display/Do not) display cumulative frequency counts/percentages. |
Options for N-way tables
table (default) | Display in table mode. |
list | Display in list mode. |
rowm (default) / norowm | [Display / Do not display] row marginals. |
colm (default) / nocolm | [Display / Do not display] column marginals. |
tabm (default) / notabm | [Display / Do not display] table marginals—only for more than two series. |
subm (default) / nosubm | [Display / Do not display] sub marginals—only for “l” option with more than two series. |
full (default) / sparse | (Full/Sparse) tabulation in list display. |
totpct / nototpct (default) | [Display / Do not display] percentages of total observations. |
tabpct / notabpct (default) | [Display / Do not display] percentages of table observations—only for more than two series. |
rowpct / norowpct (default) | [Display / Do not display] percentages of row total. |
colpct / nocolpct (default) | [Display / Do not display] percentages of column total. |
exp / noexp (default) | [Display / Do not display] expected counts under full independence. |
tabexp / notabexp (default) | [Display / Do not display] expected counts under table independence—only for more than two series. |
test (default) / notest | [Display / Do not display] tests of independence. |
Examples
matrix x(50, 4)
rndint(x, 10)
x.freq(nov, noa)
tabulates each value (no binning) of HRS in ascending order with counts, percentages, and cumulatives.
x.freq(v=200, b=100, keepna, noa)
tabulates X including NAs. The values will be binned if INC has more than 200 distinct values; EViews will create at most 100 equal value-width bins. The number of bins may be less than 100.
x.freq(v=10, norowm, nocolm)
displays tables of binned pairs of the first two columns of X for each bin/value of the remaining columns. The table will not contain row and column marginals.
x.freq(v=10, norowm, nocolm, sort=freqhilo)
displays the same table with the table rows and columns ordered from values with highest frequency to lowest.
Cross-references
See
“One-Way Tabulation” and
“N-Way Tabulation” for a discussion of frequency tables.
Imports data from a foreign file into the matrix object.
Syntax
matrix_name.import([type=]) source_description import_specification
• source_description should contain a description of the file from which the data is to be imported. The specification of the description is usually just the path and file name of the file, however you can also specify more precise information. See
wfopenfor more details on the specification of
source_description.
• The optional “type=” option may be used to specify a source type. For the most part, you should not need to specify a “type=” option as EViews will automatically determine the type from the filename. The following table summaries the various source formats with the corresponding “type=” keywords:
| |
Excel (through 2003) | “excel” |
Excel 2007 (xml) | “excelxml” |
HTML | “html” |
Text / ASCII | “text” |
• import_specification can be used to provide additional information about the file to be read. The details of import_specification will depend upon the type of file being imported.
Excel Files
The syntax for reading Excel files is:
matrix_name.import(type=excel[xml]) source_description [table_description] [variables_description]
The following table_description elements may be used when reading Excel data:
• “range = arg”, where arg is a range of cells to read from the Excel workbook, following the standard Excel format [worksheet!][topleft_cell[:bottomright_cell]].
If the worksheet name contains spaces, it should be placed in single quotes. If the worksheet name is omitted, the cell range is assumed to refer to the currently active sheet. If only a top left cell is provided, a bottom right cell will be chosen automatically to cover the range of non-empty cells adjacent to the specified top left cell. If only a sheet name is provided, the first set of non-empty cells in the top left corner of the chosen worksheet will be selected automatically. As an alternative to specifying an explicit range, a name which has been defined inside the excel workbook to refer to a range or cell may be used to specify the cells to read.
• “byrow”, transpose the incoming data. This option allows you to read files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int| all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file).
• “firstobs=int”, first observation to be imported from the data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Excel Examples
matrix_name.import "c:\data files\data.xls"
loads the active sheet of DATA.XLSX into the MATRIX_NAME matrix object.
matrix_name.import "c:\data files\data.xls" range="GDP data"
reads the data contained in the “GDP data” sheet of “Data.XLS” into the MATRIX_NAME object.
HTML Files
The syntax for reading HTML pages is:
matrix_name.import(type=html) source_description [table_description] [variables_description]
The following table_description elements may be used when reading an HTML file or page:
• “table = arg”, where arg specifies which HTML table to read in an HTML file/page containing multiple tables.
When specifying arg, you should remember that tables are named automatically following the pattern “Table01”, “Table02”, “Table03”, etc. If no table name is specified, the largest table found in the file will be chosen by default. Note that the table numbering may include trivial tables that are part of the HTML content of the file, but would not normally be considered as data tables by a person viewing the page.
• “skip = int”, where int is the number of rows to discard from the top of the HTML table.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file).
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
HTML Examples
mat1.import "c:\data.html"
loads into the MAT1 matrix object the data located in the HTML file “Data.HTML” located on the C:\ drive
mat1.import(type=html) "http://www.tradingroom.com.au/apps/mkt/forex.ac" colhead=3
loads into a matrix object MAT1 the data with the given URL located on the website site “http://www.tradingroom.com.au”. The column header is set to three rows.
Text and Binary Files
The syntax for reading text or binary files is:
matrix_name.import(type=arg) source_description [table_description] [variables_description]
If a table_description is not provided, EViews will attempt to read the file as a free-format text file. The following table_description elements may be used when reading a text or binary file:
• “ftype = [ascii|binary]” specifies whether numbers and dates in the file are stored in a human readable text (ASCII), or machine readable (Binary) form.
• “rectype = [crlf|fixed|streamed]” describes the record structure of the file:
“crlf”, each row in the output table is formed using a fixed number of lines from the file (where lines are separated by carriage return/line feed sequences). This is the default setting.
“fixed”, each row in the output table is formed using a fixed number of characters from the file (specified in “reclen= arg”). This setting is typically used for files that contain no line breaks.
“streamed”, each row in the output table is formed by reading a fixed number of fields, skipping across lines if necessary. This option is typically used for files that contain line breaks, but where the line breaks are not relevant to how rows from the data should be formed.
• “reclines =int”, number of lines to use in forming each row when “rectype=crlf” (default is 1).
• “reclen=int”, number of bytes to use in forming each row when “rectype=fixed”.
• “recfields=int”, number of fields to use in forming each row when “rectype=streamed”.
• “skip=int”, number of lines (if rectype is “crlf”) or bytes (if rectype is not “crlf”) to discard from the top of the file.
• “comment=string“, where string is a double-quoted string, specifies one or more characters to treat as a comment indicator. When a comment indicator is found, everything on the line to the right of where the comment indicator starts is ignored.
• “emptylines=[keep|drop]”, specifies whether empty lines should be ignored (“drop”), or treated as valid lines (“keep”) containing missing values. The default is to ignore empty lines.
• “tabwidth=int”, specifies the number of characters between tab stops when tabs are being replaced by spaces (default=8). Note that tabs are automatically replaced by spaces whenever they are not being treated as a field delimiter.
• “fieldtype=[delim|fixed|streamed|undivided]”, specifies the structure of fields within a record:
“Delim”, fields are separated by one or more delimiter characters
“Fixed”, each field is a fixed number of characters
“Streamed”, fields are read from left to right, with each field starting immediately after the previous field ends.
“Undivided”, read entire record as a single series.
• “quotes=[single|double|both|none]”, specifies the character used for quoting fields, where “single” is the apostrophe, “double” is the double quote character, and “both” means that either single or double quotes are allowed (default is “both”). Characters contained within quotes are never treated as delimiters.
• “singlequote“, same as “quotes = single”.
• “delim=[comma|tab|space|dblspace|white|dblwhite]”, specifies the character(s) to treat as a delimiter. “White” means that either a tab or a space is a valid delimiter. You may also use the abbreviation “d=” in place of “delim=”.
• “custom="arg1"”, specifies custom delimiter characters in the double quoted string. Use the character “t” for tab, “s” for space and “a” for any character.
• “mult=[on|off]”, to treat multiple delimiters as one. Default value is “on” if “delim” is “space”, “dblspace”, “white”, or “dblwhite”, and “off” otherwise.
• “endian = [big|little]”, selects the endianness of numeric fields contained in binary files.
• “string = [nullterm|nullpad|spacepad]”, specifies how strings are stored in binary files. If “nullterm”, strings shorter than the field width are terminated with a single zero character. If “nullpad”, strings shorter than the field width are followed by extra zero characters up to the field width. If “spacepad”, strings shorter than the field width are followed by extra space characters up to the field width.
• “byrow”, transpose the incoming data. This option allows you to import files where the series are contained in rows (one row per series) rather than columns.
• “lastcol”, include implied last column. For lines that end with a delimiter, this option adds an additional column. When importing a CSV file, lines which have the delimiter as the last character (for example: “name, description, date,”), EViews normally determines the line to have 3 columns. With the above option, EViews will determine the line to have 4 columns. Note this is not the same as a line containing “name, description, date”. In this case, EViews will always determine the line to have 3 columns regardless if the option is set.
A central component of the table_description element is the format statement. You may specify the data format using the following table descriptors:
• Fortran Format:
fformat=([n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)
where Type specifies the underlying data type, and may be one of the following,
I - integer
F - fixed precision
E - scientific
A - alphanumeric
X - skip
and n1, n2, ... are the number of times to read using the descriptor (default=1). More complicated Fortran compatible variations on this format are possible.
• Column Range Format:
rformat="[n1]Type[Width][.Precision], [n2]Type[Width][.Precision], ...)"
where optional type is “$” for string or “#” for number, and n1, n2, n3, n4, etc. are the range of columns containing the data.
• C printf/scanf Format:
cformat="fmt"
where fmt follows standard C language (printf/scanf) format rules.
The optional variables_description may be formed using the elements:
• “colhead=int”, number of table rows to be treated as column headers.
• “na="arg1"”, text used to represent observations that are missing from the file. The text should be enclosed on double quotes.
• “scan=[int|all]”, number of rows of the table to scan during automatic format detection (“scan=all” scans the entire file).
• “firstobs=int”, first observation to be imported from the table of data (default is 1). This option may be used to start reading rows from partway through the table.
• “lastobs = int”, last observation to be read from the table of data (default is last observation of the file). This option may be used to read only part of the file, which may be useful for testing.
Text and Binary File Examples (.txt, .csv, etc.)
mat2.import c:\data.csv skip=5
reads “Data.CSV” into a MAT2, skipping the first 5 rows.
mat2.import(type=text) c:\date.txt delim=comma
loads the comma delimited data “Date.TXT” into the MAT2 matrix object.
Cross-references
Display or change the label view of a matrix, including the last modified date and display name (if any).
As a procedure, label changes the fields in the matrix label.
Syntax
matrix_name.label
matrix_name.label(options) [text]
Options
The first version of the command displays the label view of the matrix. The second version may be used to modify the label. Specify one of the following options along with optional text. If there is no text provided, the specified field will be cleared.
c | Clears all text fields in the label. |
d | Sets the description field to text. |
s | Sets the source field to text. |
u | Sets the units field to text. |
r | Appends text to the remarks field as an additional line. |
p | Print the label view. |
Examples
The following lines replace the remarks field of M1 with “Data from CPS 1988 March File”:
m1.label(r)
m1.label(r) Data from CPS 1988 March File
To append additional remarks to M1, and then to print the label view:
m1.label(r) Log of hourly wage
m1.label(p)
To clear and then set the units field, use:
m1.label(u) Millions of bushels
Cross-references
See
“Labeling Objects” for a discussion of labels.
Save the scores from a principal components analysis of the series in a matrix.
Syntax
matrix_name.makepcomp(options) output_list
where the
output_list is a list of names identifying the saved components. EViews will save the first
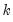
components corresponding to the
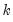
elements in
output_list, up to the total number of series in the group.
Options
scale=arg (default=“normload”) | Diagonal matrix scaling of the loadings and the scores: normalize loadings (“normload”), normalize scores (“normscores”), symmetric weighting (“symmetric”), user-specified (arg=number). |
cpnorm | Compute the normalization for the score so that cross-products match the target (by default, EViews chooses a normalization scale so that the moments of the scores match the target). |
eigval=vec_name | Specify name of vector to hold the saved the eigenvalues in workfile. |
eigvec=mat_name | Specify name of matrix to hold the save the eigenvectors in workfile. |
prompt | Force the dialog to appear from within a program. |
Covariance Options
cov=arg (default=“corr”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), uncentered ordinary correlation (“ucorr”). Note that Kendall’s tau measures are not valid methods. |
wgt=name (optional) | Name of vector containing weights. The number of rows of the weight vector should match the number of rows in the original matrix. |
wgtmethod=arg (default = “sstdev” | Weighting method: frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations where “weights=” is specified. Weights for rank correlation and Kendall’s tau calculations are always frequency weights. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction accounting for the mean (for centered specifications) and any partial conditioning variables. The default behavior in these cases is to perform no adjustment ( e.g. – compute sample covariance dividing by 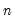 rather than 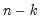 ). |
Examples
mat1.makepcomp comp1 comp2 comp3
saves the first three principal components (in normalized loadings form) to the workfile. The components will have variances that are proportional to the eigenvalues.
mat1.makepcomp(scale=normscore) comp1 comp2 comp3
normalizes the scores so that the resulting series have variances that are equal to 1.
You may change the scaling for the normalized components so that the cross-products equal 1, using the cpnorm option:
mat1.makepcomp(scale=normscore, cpnorm) comp1 comp2 comp3
Cross-references
See
“Saving Component Scores” for further discussion. See
Matrix::pcomp for tools to display the principal components results for the matrix.
Declare and optionally initializes a matrix object.
Syntax
matrix(r, c) matrix_name[=assignment]
The matrix keyword is followed by the name you wish to give the matrix. matrix also takes an optional argument specifying the row r and column c dimension of the matrix. Once declared, matrices may be resized by repeating the matrix command using the original name.
You may combine matrix declaration and assignment. If there is no assignment statement, the matrix will initially be filled with zeros.
You should use sym for symmetric matrices.
Examples
matrix mom
declares a matrix named MOM with one element, initialized to zero.
matrix(3,6) coefs
declares a 3 by 6 matrix named COEFS, filled with zeros.
Cross-references
See
“Matrix Language” for further discussion.
See
“Rowvector” and
“Vector” and
“Sym” for full descriptions of the various matrix objects.
Push updates to OLE linked objects in open applications.
Syntax
matrix_name.olepush
Cross-references
See
“Object Linking and Embedding (OLE)” for a discussion of using OLE with EViews.
Principal components analysis of the columns in a matrix.
Syntax
There are two forms of the pcomp command. The first form, which applies when displaying eigenvalue table output or graphs of the ordered eigenvalues, has only options and no command argument.
matrix_name.pcomp(options)
The second form, which applies to the graphs of component loadings, component scores, and biplots, uses the optional argument to determine which components to plot. In this form:
matrix_name.pcomp(options) [graph_list]
where the [graph_list] is an optional list of integers and/or vectors containing integers identifying the components to plot. Multiple pairs are handled using the method specified in the “mult=” option.
If the list of component indices omitted, EViews will plot only first and second components. Note that the order of elements in the list matters; reversing the order of two indices reverses the axis on which each component is displayed.
Options
out=arg (default=“table”) | Output: table of eigenvalue and eigenvector results (“table”), graphs of ordered eigenvalues (“graph”), graph of the eigenvectors (“loadings”), graph of the component scores (“scores”), biplot of the loadings and scores (“biplot”). Note: when specifying the eigenvalue graph (“out=graph”), the option keywords “scree” (scree graph), “diff” (difference in successive eigenvalues), and “cproport” (cumulative proportion of total variance) may be included to control the output. By default, EViews will display the scree graph. If you may one or more the three keywords, EViews will construct the graph using only the specified types. |
eigval=vec_name | Specify name of vector to hold the saved the eigenvalues in workfile. |
eigvec=mat_name | Specify name of matrix to hold the save the eigenvectors in workfile. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Number of Component Options
fsmethod=arg (default=“simple”) | Component retention method: “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), “simple” (simple eigenvalue methods), “user” (user-specified value). Note the following: (1) If using simple methods, the minimum eigenvalue and cumulative proportions may be specified using “minigen=” and “cproport=”. (2) If setting “fsmethod=user” to provide a user-specified value, you must specify the value with “r=”. |
r=arg (default=1) | User-specified number of components to retain (for use when “fsmethod=user”). |
mineigen=arg (default=0) | Minimum eigenvalue to retain component (when “fsmethod=simple”). |
cproport=arg (default=1.0) | Cumulative proportion of eigenvalue total to attain (when “fsmethod=simple”). |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 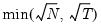 ), “size” ( 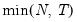 ) ), “user” (user specified value), where 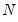 is the number of columns of the matrix and 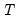 is the number of rows. (1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let for 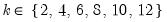 and let and let 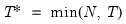 ; then the default maximum lag is given by ; then the default maximum lag is given by |
n=arg or rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Component selection criterion when “fsmethod=bn”: “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Component selection criterion when “fsmethod=ah”: “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Component selection when “fsmethod=simple”: “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures. |
sdizetime | Standardizes observations across time prior to component selection procedures. |
demeancross | Demeans observations across cross-sections prior to component selection procedures. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures. |
Covariance Options
cov=arg (default=“cov”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), Kendall’s tau-b (“taub”), Kendall’s tau-a (“taua”), uncentered ordinary covariance (“ucov”), uncentered ordinary correlation (“ucorr”). |
wgt=name (optional) | Name of vector containing weights. The number of rows of the weight vector should match the number of rows in the original matrix. |
wgtmethod=arg (default = “sstdev” | Weighting method: frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations where “weights=” is specified. Weights for rank correlation and Kendall’s tau calculations are always frequency weights. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction accounting for the mean (for centered specifications) and any partial conditioning variables. The default behavior in these cases is to perform no adjustment ( e.g. – compute sample covariance dividing by 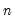 rather than 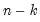 ). |
Graph Options
scale=arg, (default=“normload”) | Diagonal matrix scaling of the loadings and the scores: normalize loadings (“normload”), normalize scores (“normscores”), symmetric weighting (“symmetric”), user-specified (arg=number). |
mult =arg (default=“first”) | Multiple series handling: plot first against remainder (“first”), plot as x-y pairs (“pair”), lower-triangular plot (“lt”). |
nocenter | Do not center graphs around the origin. By default, EViews centers biplots around (0, 0). |
labels=arg, (default=“outlier”) | Observation labels for the scores: outliers only (“outlier”), all points (“all”), none (“none”). |
labelprob=number | Probability value for determining whether a point is an outlier according to the chi-square tests based on the squared Mahalanbois distance between the observation and the sample means (when using the “labels=outlier” option). |
autoscale=arg | Scale factor applied to the automatically specified loadings when displaying both loadings and scores). The default is to let EViews auto-choose a scale or to specify “userscale=” to scale the original loadings. |
userscale=arg | Scale factor applied to the original loadings when displaying both loadings and scores). The default is to let EViews auto-choose a scale, or to specify “autoscale=” to scale the automatically scaled loadings. |
cpnorm | Compute the normalization for the score so that cross-products match the target (by default, EViews chooses a normalization scale so that the moments of the scores match the target). |
Examples
freeze(tab1) mat1.pcomp(method=corr, eigval=v1, eigvec=m1)
stores the table view of the eigenvalues and eigenvectors of MAT1 in a table object named tab1, the eigenvalues in a vector named v1, and the eigenvectors in a matrix named m1.
mat1.pcomp(method=cov, out=graph)
displays the scree plot of the ordered eigenvalues computed from the covariance matrix.
mat1.pcomp(method=rcorr, out=biplot, scale=normscores)
displays a biplot where the scores are normalized to have variances that equal the eigenvalues of the Spearman correlation matrix computed for the series in MAT1.
Cross-references
See
“Principal Components” for further discussion. See also
“Covariance Analysis” for discussion of the preliminary computation.
Note that this view analyzes the eigenvalues and eigenvectors of a covariance (or other association) matrix computed from the series in a group or the columns of a matrix. You may use
Sym::eigen to examine the eigenvalues of a symmetric matrix.
Import data from a foreign disk file into a matrix.
(This is a deprecated method of importing into a matrix. See
Matrix::import for the currently supported method.)
May be used to import data into an existing workfile from a text, Excel, or Lotus file on disk.
Syntax
matrix_name.read(options) [path\]file_name
You must supply the name of the source file. If you do not include the optional path specification, EViews will look for the file in the default directory. Path specifications may point to local or network drives. If the path specification contains a space, you may enclose the entire expression in double quotation marks.
Options
File type options
t=dat, txt | ASCII (plain text) files. |
t=wk1, wk3 | Lotus spreadsheet files. |
t=xls | Excel spreadsheet files. |
If you do not specify the “t” option, EViews uses the file name extension to determine the file type. If you specify the “t” option, the file name extension will not be used to determine the file type.
Options for ASCII text files
t | Read data organized by column (transposed). Default is to read by row. |
na=text | Specify text for NAs. Default is “NA”. |
d=t | Treat tab as delimiter (note: you may specify multiple delimiter options). The default is “d=c” only. |
d=c | Treat comma as delimiter. |
d=s | Treat space as delimiter. |
d=a | Treat alpha numeric characters as delimiter. |
custom = symbol | Specify symbol/character to treat as delimiter. |
mult | Treat multiple delimiters as one. |
rect (default) / norect | [Treat / Do not treat] file layout as rectangular. |
skipcol = integer | Number of columns to skip. Must be used with the “rect” option. |
skiprow = integer | Number of rows to skip. Must be used with the “rect” option. |
comment= symbol | Specify character/symbol to treat as comment sign. Everything to the right of the comment sign is ignored. Must be used with the “rect” option. |
singlequote | Strings are in single quotes, not double quotes. |
dropstrings | Do not treat strings as NA; simply drop them. |
negparen | Treat numbers in parentheses as negative numbers. |
allowcomma | Allow commas in numbers (note that using commas as a delimiter takes precedence over this option). |
Options for spreadsheet (Lotus, Excel) files
t | Read data organized by column (transposed). Default is to read by row. |
letter_number (default=“b2”) | Coordinate of the upper-left cell containing data. |
s=sheet_name | Sheet name for Excel 5–8 Workbooks. |
Examples
m1.read(t=dat,na=.) a:\mydat.raw
reads data into matrix M1 from an ASCII file MYDAT.RAW in the A: drive. The data in the file are listed by row, and the missing value NA is coded as a “.” (dot or period).
m1.read(t,a2,s=sheet3) cps88.xls
reads data into matrix M1 from an Excel file CPS88 in the default directory. The data are organized by column (transposed), the upper left data cell is A2, and the data is read from a sheet named SHEET3.
m2.read(a2, s=sheet2) "\\network\dr 1\cps91.xls"
reads the Excel file CPS91 into matrix M2 from the network drive specified in the path.
Cross-references
See
“Importing Data” for a discussion and examples of importing data from external files.
Resample from rows in a group.
Syntax
group_name.resample(options) output_name
You should follow the resample keyword and options with an output_name containing a name of the matrix to hold the output.
By default, EViews uses the output_name “*_b” so that “_b” will be appended to the name of the original matrix and used as the output name.
Options
rows = n | (Optional) Number of rows of the output matrix. Default is to create an output matrix with the same the number of rows as the source matrix. |
permute | Draw from rows without replacement. Default is to draw with replacement. |
weight= vector_name | Name of vector to be used for weighted sampling, containing values proportional to the desired row probabilities (importance sampling). The weight vector must have the same number of rows as the matrix, with non-missing, non-negative values. The weight values need not add up to 1, as EViews will normalize the weights. If no weights are provided, rows will be drawn with equal probability weights. |
block=integer | Block length for each draw. Must be a positive integer. The default block length is 1. |
withna (default) | [Draw / Do not draw] from all rows in the matrix, including those with NAs. |
dropna | Do not draw from rows that contain missing values in the matrix. |
fixna | Excludes NAs from draws but copies rows containing missing values to the output matrix. |
prompt | Force the dialog to appear from within a program. |
• Block bootstrap (“block=” length larger than 1) requires a contiguous output. Therefore a block length larger than 1 cannot be used together with the “fixna” option.
• The “fixna” option will have an effect only if there are missing values in the rows of the source matrix.
• If you specify “fixna”, we first copy rows with any missing values in the matrix sample to the output matrix (if relevant). Then the remaining rows of the output matrix will be resampled from the remaining rows of the input matrix.
• If you choose “dropna” and the block length is larger than 1, the rows of the matrix used for simulation may adjust in order to ensure that there are no missing values in any of the drawn blocks.
• If you choose “permute”, the block option will be reset to 1, the “dropna” and “fixna” options will be ignored (reset to the default “withna” option), and the “weight” option will be ignored (reset to default equal weights).
Examples
matrix(100, 5) f
nrnd(f)
f.resample(n = 1000) f_resample
creates a new matrix F_RESAMPLE obtained by drawing 1000 rows with replacement from F.
f.resample(weight=wt) f_weighted
will compute a weighted resample and save the results to the same sized matrix F_WEIGHTED. The rows in the source will be drawn with probabilities proportional to the corresponding values in the series WT. WT must have the same number of rows as F and must contain non-missing, non-negative values.
Cross-references
See
“Resample” for a discussion of the resampling procedure. For additional discussion of wildcards, see
Appendix A. “Wildcards”.
See also
@resample and
@permute for sampling from matrices.
Resize the matrix object.
Syntax
matrix_name.resize rows cols
Examples
mat1.resize 3 5
resizes the matrix MAT1 to 3 rows and 5 columns, retaining the contents of any existing elements and initializing new elements to 0.
Set the object attribute.
Syntax
alpha_name.setattr(attr) attr_value
Sets the attribute attr to attr_value. Note that quoting the arguments may be required. Once added to an object, the attribute may be extracted using the @attr data member.
Examples
a.setattr(revised) never
String s = a.@attr("revised")
sets the “revised” attribute in the object A to the string “never”, and extracts the attribute into the string object S.
Cross-references
Set the column labels in a matrix object.
Syntax
matrix_name.setcollabels label1 label2 label3....
Follow the setcollabels command with a space delimited list of column labels. Note that each column label should not contain spaces unless it is enclosed in quotes. If you provide fewer labels than there are columns, EViews will keep the corresponding default column names (“C11”, “C12”, etc...).
Examples
mat1.setcollabels USA UK FRANCE
sets the column label for the first column in matrix MAT1 to USA, the second to UK, and the third to FRANCE.
Cross-references
Set the display format for cells in a matrix object spreadsheet view.
Syntax
matrix_name.setformat format_arg
where format_arg is a set of arguments used to specify format settings. If necessary, you should enclose the format_arg in double quotes.
For matrices, setformat operates on all of the cells in the matrix.
To format numeric values, you should use one of the following format specifications:
g[.precision] | significant digits |
f[.precision] | fixed decimal places |
c[.precision] | fixed characters |
e[.precision] | scientific/float |
p[.precision] | percentage |
r[.precision] | fraction |
To specify a format that groups digits into thousands using a comma separator, place a “t” after the format character. For example, to obtain a fixed number of decimal places with commas used to separate thousands, use “ft[.precision]”.
To use the period character to separate thousands and commas to denote decimal places, use “..” (two periods) when specifying the precision. For example, to obtain a fixed number of characters with a period used to separate thousands, use “ct[..precision]”.
If you wish to display negative numbers surrounded by parentheses (i.e., display the number -37.2 as “(37.2)”), you should enclose the format string in “()” (e.g., “f(.8)”).
Examples
To set the format for all cells in the matrix to fixed 5-digit precision, simply provide the format specification:
matrix1.setformat f.5
Other format specifications include:
matrix1.setformat f(.7)
matrix1.setformat e.5
Cross-references
See
Matrix::setwidth,
Matrix::setindent and
Matrix::setjust for details on setting spreadsheet widths, indentation and justification.
Set the display indentation for cells in a matrix object spreadsheet view.
Syntax
matrix_name.setindent indent_arg
where indent_arg is an indent value specified in 1/5 of a width unit. The width unit is computed from representative characters in the default font for the current spreadsheet (the EViews spreadsheet default font at the time the spreadsheet was created), and corresponds roughly to a single character. Indentation is only relevant for non-center justified cells.
The default value is taken from the Global Defaults at the time the spreadsheet view is created.
For matrices, setindent operates on all of the cells in the matrix.
Examples
To set the indentation for all the cells in a matrix object:
matrix1.setindent 2
Cross-references
See
Matrix::setwidth and
Matrix::setjust for details on setting spreadsheet widths and justification.
Set the horizontal justification for all cells in the spreadsheet view of the matrix object.
Syntax
matrix_name.setjust format_arg
where
format_arg may be set to left, center, right, or auto (strings are left-justified and numbers are right-justified). Default display settings can be set in General Options; see
“Spreadsheet Data Display”.
Examples
mat1.setjust left
left-justifies the cells in the spreadsheet view of the matrix MAT1.
Cross-references
See
Matrix::setwidth and
Matrix::setindent for details on setting spreadsheet widths and indentation.
Set the row labels in a matrix object.
Syntax
matrix_name.setrowlabels label1 label2 label3....
Follow the setrowlabels command with a space delimited list of row labels. Note that each row label should not contain spaces unless it is enclosed in quotes. If you provide fewer labels than there are rows, EViews will use the corresponding default row names (“R11”, “R12”, etc...).
Examples
mat1.setrowlabels USA UK FRANCE
sets the row label for the first row in matrix MAT1 to USA, the second to UK, and the third to FRANCE.
Cross-references
Set the column width for all columns in a matrix object spreadsheet.
Syntax
matrix_name.setwidth width_arg
where width_arg specifies the width unit value. The width unit is computed from representative characters in the default font for the current spreadsheet (the EViews spreadsheet default font at the time the spreadsheet was created), and corresponds roughly to a single character. width_arg values may be non-integer values with resolution up to 1/10 of a width unit.
Examples
mat1.setwidth 12
sets the width of all columns in matrix MAT1 to 12 width units.
Cross-references
See
Matrix::setindent and
Matrix::setjust for details on setting spreadsheet indentation and justification.
Spreadsheet view of a matrix object.
Syntax
matrix_name.sheet(options)
Options
p | Print the spreadsheet view. |
Examples
mat1.sheet(p)
displays and prints the spreadsheet view of matrix MAT1.
Displays the custom row and column labels of a matrix spreadsheet.
Syntax
matrix_name.showlabels mode
where mode is either 0 or 1 where 0 displays the default row and column labels and 1 displays the custom row and column labels (if present).
Examples
m1.showlabels 1
displays the custom row and column labels for the M1 spreadsheet. If custom labels have not been set the default labels will be displayed.
m1.showlabels 0
displays the default row and column labels for the M1 spreadsheet.
Cross-references
Descriptive statistics for columns of a matrix.
Computes and displays a table of means, medians, maximum and minimum values, standard deviations, and other descriptive statistics for each column of the matrix.
Syntax
matrix_name.stats(options)
Options
i | Individual sample for each column after removing missing values for the column. By default, EViews computes the statistics using a common sample after removing rows with missing values. |
p | Print the stats table. |
Examples
The command
matrix1.stats
computes the descriptive statistics for each column of MATRIX1 after listwise deletion of rows with missing values. Alternately,
matrix1.stats(i)
displays the descriptive statistics view of matrix1 showing the statistics for each column of the matrix computed using individual samples.
Cross-references
See
“Descriptive Statistics” for a discussion of the descriptive statistics views of a group.
Test equality of the mean, median or variance of the series in the group.
Syntax
group_name.testbtw(options)
By default, testbtw will test for equality of means, but you may specify instead tests of medians or variances as an option, and choose whether to use balanced or unbalanced samples.
Options
mean (default) | Test equality of mean. |
med | Test equality of median. |
var | Test equality of variance. |
c | Use common (balanced) sample for computing statistics after removing observations with missing values for any series in the group. By default, EViews computes the statistics for each series using an individual (potentially unbalanced) sample after removing observations with missing values for the series. |
prompt | Force the dialog to appear from within a program. |
p | Print the test results. |
Examples
The commands
group g1 wage_m wage_f
g1.testbtw
uses both parametric ANOVA and nonparametric approaches to test the equality of means of the two series WAGE_M and WAGE_F using individual samples to compute the means. Alternately, the command,
g1.testbtw(var, c)
tests for equality of variances using a balanced sample after listwise deletion of rows with missing values.
Cross-references
See
“Tests of Equality” for further discussion of these tests.
Test equality of the mean, median or variances of the columns in a matrix.
Syntax
matrix_name.testbtw(options)
By default, testbtw will test for equality of means, but you may specify instead tests of medians or variances as an option, and choose whether to use balanced or unbalanced samples.
Options
mean (default) | Test equality of means. |
med | Test equality of medians. |
var | Test equality of variances. |
c | Use common (balanced) sample for computing statistics after removing rows with missing values for any element in the row. By default, EViews computes the statistics for each column using a individual (potentially unbalanced) sample after removing observations with missing values for the column. |
prompt | Force the dialog to appear from within a program. |
p | Print the test results. |
Examples
The command
mat1.testbtw
uses both parametric ANOVA and nonparametric approaches to test the equality of the means of the columns of MAT1 using individual samples. Alternately, the command,
mat1.testbtw(var, c)
tests for equality of the column variances using a balanced sample after listwise deletion of rows with missing values.
Cross-references
See
“Tests of Equality” for further discussion of these tests.
Write EViews data to a text (ASCII), Excel, or Lotus file on disk.
Creates a foreign format disk file containing EViews data. May be used to export EViews data to another program.
This routine should realistically only be used in the oft-hand chance that you wish to write into a Lotus file. Improved Excel, text, and other format writing is available in
Matrix::export.
Syntax
matrix_name.write(options) [path\filename]
Follow the name of the matrix object by a period, the keyword, and the name for the output file. The optional path name may be on the local machine, or may point to a network drive. If the path name contains spaces, enclose the entire expression in double quotation marks. The entire matrix will be exported.
Note that EViews cannot, at present, write into an existing file. The file that you select will, if it exists, be replaced.
Options
Options are specified in parentheses after the keyword and are used to specify the format of the output file.
File type
t=dat, txt | ASCII (plain text) files. |
t=wk1, wk3 | Lotus spreadsheet files. |
t=xls | Excel spreadsheet files. |
If you omit the “t=” option, EViews will determine the type based on the file extension. Unrecognized extensions will be treated as ASCII files. For Lotus and Excel spreadsheet files specified without the “t=” option, EViews will automatically append the appropriate extension if it is not otherwise specified.
ASCII text files
na=string | Specify text string for NAs. Default is “NA”. |
d=arg | Specify delimiter (default is tab): “s” (space), “c” (comma). |
t | Write by column (transpose the data). Default is to write by row. |
Spreadsheet (Lotus, Excel) files
letter_number | Coordinate of the upper-left cell containing data. |
t | Write by column (transpose the data). Default is to write by row. |
Examples
m1.write(t=txt,na=.) a:\dat1.csv
Writes the matrix M1 into an ASCII file named DAT1.CSV on the A: drive. NAs are coded as “.” (dot).
m1.write(t=txt,na=.) dat1.csv
writes the same file in the default directory.
m1.write(t=xls) "\\network\drive a\results"
saves the contents of M1 in an Excel file “Results.xls” in the specified directory.
Cross-references
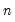 -way contingency table.
-way contingency table.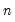 -way contingency table.
-way contingency table.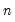 -way contingency table.
-way contingency table.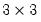 Pearson correlation matrix for the columns series in MAT1.
Pearson correlation matrix for the columns series in MAT1.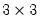 Pearson covariance matrix for the columns series in MAT1.
Pearson covariance matrix for the columns series in MAT1.
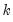 components corresponding to the
components corresponding to the 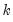 elements in output_list, up to the total number of series in the group.
elements in output_list, up to the total number of series in the group.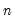 rather than
rather than 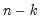 ).
).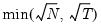 ), “size” (
), “size” (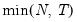 ), “user” (user specified value), where
), “user” (user specified value), where 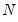 is the number of columns of the matrix and
is the number of columns of the matrix and 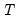 is the number of rows.
is the number of rows.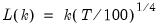
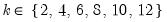 and let
and let 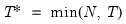 ; then the default maximum lag is given by
; then the default maximum lag is given by 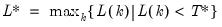
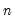 rather than
rather than 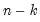 ).
).