System
System of equations for estimation.
System Declaration
Declare a system object by entering the keyword system, followed by a name:
system mysys
To fill a system, open the system and edit the specification view, or use
append. Note that systems are not used for simulation. See
“Model”.
System Methods
3sls three-stage least squares.
arch estimate generalized autoregressive conditional heteroskedasticity (GARCH) models.
fiml full information maximum likelihood.
gmm generalized method of moments.
ls ordinary least squares.
sur seemingly unrelated regression.
tsls two-stage least squares.
wls weighted least squares.
wtsls weighted two-stage least squares.
System Views
cellipse confidence ellipses for coefficient restrictions.
coefcov coefficient covariance matrix.
correl display graphs or tables of residual autocorrelations and cross-correlations.
derivs derivatives of the system equations.
display display table, graph, or spool in object window.
endog table or graph of endogenous variables.
estcov display the covariance matrix used in estimation.
garch conditional variance/covariance of (G)ARCH estimation.
grads examine the gradients of the objective function.
jbera multivariate residual normality test.
label label information for the system object.
output table of estimation results.
qstats multivariate residual autocorrelation Portmanteau tests.
resids residual graphs or spreadsheets.
results table of estimation results.
spec text representation of system specification.
wald Wald coefficient restriction test.
System Procs
append add a line of text to the system specification.
autospec automatically create system specification text.
clearhist clear the contents of the history attribute.
copy creates a copy of the system.
makegarch generate conditional variance series.
makeloglike create and save log likelihood contribution from system (ARCH estimation).
makemodel create a model from the estimated system.
makeresids make series containing residuals from system.
olepush push updates to OLE linked objects in open applications.
setattr set the value of an object attribute.
System Data Members
Scalar Values (individual equation data)
@coefcov(i, j) covariance of coefficients i and j.
@coefs(i) coefficient i.
@dw(k) Durbin-Watson statistic for equation k.
@eqncoef(k) number of estimated coefficients in equation k.
@eqregobs(k) number of observations in equation k.
@meandep(k) mean of the dependent variable in equation k.
@r2(k) R-squared statistic for equation k.
@rbar2(k) adjusted R-squared statistic for equation k.
@sddep(k) standard deviation of dependent variable in equation k.
@se(k) standard error of the regression in equation k.
@ssr(k) sum of squared residuals in equation k.
@stderrs(i) standard error for coefficient i.
@tstats(i) t-statistic or z-statistic for coefficient i.
c(i) i-th element of default coefficient vector for system (if applicable).
Scalar Values (system level data)
@aic Akaike information criterion for the system (if applicable).
@detresid determinant of the residual covariance matrix.
@hq Hannan-Quinn information criterion for the system (if applicable).
@jstat J-statistic — value of the GMM objective function (for GMM estimation).
@linecount scalar containing the number of lines in the System object.
@logl value of the log likelihood function for the system (if applicable).
@ncoefs total number of estimated coefficients in system.
@neqn number of equations.
@regobs number of observations in the sample range used for estimation (“@regobs” will differ from “@eqregobs” if the unbalanced sample is non-overlapping).
@schwarz Schwarz information criterion for the system (if applicable).
@totalobs sum of “@eqregobs” from each equation.
Vectors and Matrices
@coefcov covariance matrix for coefficients of equation.
@coefs coefficient vector.
@estcov (sym) residual covariance matrix used in estimation (see
System::estcov).
@pvals vector containing the coefficient probability values.
@residcov (sym) covariance matrix of the residuals.
@stderrs vector of standard errors for coefficients.
@tstats vector of t-statistic or z-statistic values for coefficients.
String values
@attr("arg") string containing the value of the arg attribute, where the argument is specified as a quoted string.
@command full command line form of the estimation command. Note this is a combination of @method and @options.
@description string containing the System object’s description (if available).
@detailedtype returns a string with the object type: “SYSTEM”.
@displayname returns the System’s display name. If the System has no display name set, the name is returned.
@line(i) returns a string containing the i-th line of the System object.
@method command line form of estimation method type (“ARCH”, “LS”, etc....).
@name returns the System’s name.
@options command line form of estimation options.
@remarks string containing the system object’s remarks (if available).
@smpl sample used for estimation.
@svector returns an Svector where each element is a line of the System object.
@svectornb same as @svector, with blank lines removed.
@type returns a string with the object type: “SYSTEM”.
@updatetime returns a string representation of the time and date at which the System was last updated.
System Examples
To estimate a system using GMM and to create residual series for the estimated system:
sys1.gmm(i,m=7,c=.01,b=v)
sys1.makeresids consres incres saveres
To test coefficients using a Wald test:
sys1.wald c(1)=c(4)
To save the coefficient covariance matrix:
sym covs=sys1.@coefcov
Estimate a system of equations by three-stage least squares.
Syntax
system_name.3sls(options)
Options
i | Iterate simultaneously over the weighting matrix and coefficient vector. |
s | Iterate sequentially over the weighting matrix and coefficient vector. |
o (default) | Iterate the coefficient vector to convergence following one-iteration of the weighting matrix. |
c | One step (iteration) of the coefficient vector following one-iteration of the weighting matrix. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage coefficient estimation to get the one-step weighting matrix. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
sys1.3sls(i)
Estimates SYS1 by the 3SLS method, iterating simultaneously on the weighting matrix and the coefficient vector.
nlsys.3sls(showopts,m=500)
Estimates NLSYS by 3SLS with up to 500 iterations. The “showopts” option displays the starting values and other estimation options.
Cross-references
See
“System Estimation” for discussion of system estimation.
Append a specification line to a system.
Syntax
system_name.append text
Type the text to be added after the append keyword.
Examples
system macro1
macro1.append cons=c(1)+c(2)*gdp+c(3)*cons(-1)
macro1.append inv=c(4)+c(5)*tb3+c(6)*d(gdp)
macro1.append gdp=cons+inv+gov
macro1.append inst tb3 gov cons(-1) gdp(-1)
macro1.gmm
show macro1.results
The first line declares a system. The next three lines append the specification of each endogenous variable in the system. The fifth line appends the list of instruments to be used in estimation. The last two lines estimate the model by GMM and display the estimation results.
Cross-references
Estimate generalized autoregressive conditional heteroskedasticity (GARCH) models.
Syntax
For a Diagonal VECH model:
system_name.arch(options) @diagvech c(arg) [arch(n, arg)] [tarch(n, arg)] [garch(n, arg)] [exog(series, arg)]
Indicate a Diagonal VECH model by using the @diagvech keyword. Follow the keyword with the constant term, c, and other optional terms to include in the variance equation: arch, garch, tarch, or exog (exogenous variable).
n indicates the order of the term, and arg indicates the type of coefficient for the term. For the exogenous variable, series indicates a series name.
Diagonal VECH Argument Options
c(arg) | where arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, “indef” (indefinite - default), or “vt” (variance target). |
arch(n, arg) | where n indicates the order of the term, and arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, or “indef” (indefinite - default). |
garch(n, arg) | where n indicates the order of the term, and arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, or “indef” (indefinite - default). |
tarch(n, arg) | where n indicates the order of the term, and arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, or “indef” (indefinite - default). |
exog(series, arg) | where series indicates a series name, and arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, or “indef” (indefinite - default). |
For example, “c(indef)” instructs EViews to use an indefinite matrix for the constant term, while “ARCH(1, fullrank)” includes a first order ARCH with a full rank matrix coefficient type.
For a Constant Conditional Correlation model:
system_name.arch(options) @ccc c(arg) [arch(n[, arg])] [tarch(n[, arg])] [garch(n[, arg])] [exog(series, arg)]
Indicate a Constant Conditional Correlation model by using the @ccc keyword. Follow the keyword with the constant term, c, and other optional terms to include in the variance equation: arch, garch, tarch, or exog (exogenous variable).
n indicates the order of the term, and arg indicates the type of coefficient for the term. For the exogenous variable, series indicates a series name.
Constant Conditional Correlation Argument Options
c(arg) | where arg may be “scalar” (default) or “vt” (variance target). |
arch(n[, arg]) | where n indicates the order of the term, and the optional arg may be “scalar” (default). |
garch(n[, arg]) | where n indicates the order of the term, and the optional arg may be “scalar” (default). |
tarch(n[, arg]) | where n indicates the order of the term, and the optional arg may be “scalar” (default). |
exog(series, arg) | where series indicates a series name, and arg may be “indiv” (individual - default) or “common”. |
For a Diagonal BEKK model:
system_name.arch(options) @diagbekk c(arg) [arch(n[, arg])] [tarch(n[, arg])] [garch(n[, arg])] [exog(series, arg)]
Indicate a Diagonal BEKK model by using the @diagbekk keyword. Follow the keyword with the constant term, c, and other optional terms to include in the variance equation: arch, garch, tarch, or exog (exogenous variable).
n indicates the order of the term, and arg indicates the type of coefficient for the term. For the exogenous variable, series indicates a series name.
Diagonal BEKK Argument Options
c(arg) | where arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, “indef” (indefinite - default), or “vt” (variance target). |
arch(n[, arg]) | where n indicates the order of the term, and the optional arg may be “diag” (diagonal - default). |
garch(n[, arg]) | where n indicates the order of the term, and the optional arg may be “diag” (diagonal - default). |
tarch(n[, arg]) | where n indicates the order of the term, and the optional arg may be “diag” (diagonal - default). |
exog(series, arg) | where series indicates a series name, and arg may be “scalar”, “diag” (diagonal), “rank1” (rank one), “fullrank”, or “indef” (indefinite - default). |
Options
General Options
tdist | Estimate the model assuming that the residuals follow a conditional Student’s t-distribution (the default is the conditional normal distribution). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). “bfgs” is the default for new equations. |
optstep = arg | Step method: “marquardt” (Marquardt - default); “dogleg” (Dogleg); “linesearch” (Line search). (Applicable when “optmethod=bfgs”, “optmethod=newton” or “optmethod=opg”.) |
b | Use Berndt-Hall-Hall-Hausman (BHHH) as maximization algorithm. The default is Marquardt. (Applicable when “optmethod=legacy”.) |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “bollerslev” (Bollerslev-Wooldridge method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “ (Applicable when non-legacy “optmethod=” with “cov=ordinary”.) |
h | Bollerslev-Wooldridge robust quasi-maximum likelihood (QML) covariance/standard errors. (Applicable for “optmethod=legacy” when estimating assuming normal errors.) |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector of a system’s variance component; the default behavior is to use the “C” coefficient vector. |
backcast=n | Backcast weight to calculate value used as the presample conditional variance. Weight needs to be greater than 0 and less than or equal to 1; the default value is 0.7. Note that a weight of 1 is equivalent to no backcasting, i.e. using the unconditional residual variance as the presample conditional variance. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
system sys01
sys01.append dlog(jy)=c(1)
sys01.append dlog(bp)=c(2)
sys01.arch @diagvech c(indef) arch(1,indef) garch(1,rank1)
creates a system SYS01, appends two equations, and estimates the system using maximum likelihood with ARCH. A Diagonal VECH model is used with the constant and order 1 ARCH coefficient matrix indefinite and order 1 GARCH coefficient rank 1 matrix.
sys01.arch @diagbekk c(fullrank) arch(1) garch(1)
estimates SYS01 using a Diagonal BEKK model of order (1,1), with constant coefficient a full rank matrix.
sys01.arch(backcast=1) @ccc c arch(1) garch(1) exog(x1,indiv) exog(x2,common)
estimates a CCC model, with each variance equation GARCH(1,1) and two exogenous variables X1 and X2. The influence of X1 on each variance equation can be varying, while X2’s coefficient is the same across all variance equations. Presample uses the unconditional variance since the backcast parameter is set to one.
Cross-references
See
“ARCH and GARCH Estimation” for a discussion of ARCH models.
Automatically create system specification text.
Syntax
system_name.autospec(options) y1 y2 y3 ... @reg x1 x2 x3 ... [@eqreg w1 w2 ...] [@inst z1 z2 ...] [@eqinst z3 z4 ...]
Defines the specification of the system. The @reg list consists of regressors with common coefficients in the system. The @eqreg list consists of regressors with different coefficients in each equation. The list of variables that follow @inst are the common instruments. The list of variables that follow @eqinst are the equation specific instruments.
Options
ytrans=arg | Dependent variable transformation: none (default), log (“log”), difference (“d”), difference of logs (“dlog”), one percentage change in decimal (“pch”), one-period percentage change—annualized, in percent (“pcha”), one-year percentage change in decimal (“pchy”). |
prompt | Force the dialog to appear from within a program. |
Examples
system sys1
sys1.autospec @regs y1 y2 y3 @regs x1 x2 c @inst z1 z2 z3
creates a system named SYS1 with the series Y1, Y2 and Y3 as the dependent variables and a common intercept and coefficients on X1 and X2, with common instruments Z1, Z2, and Z3.
Cross-references
See
“System Estimation” for a discussion of system objects in EViews.
Confidence ellipses for coefficient restrictions.
The cellipse view displays confidence ellipses for pairs of coefficient restrictions for an estimation object.
Syntax
system_name.cellipse(options) restrictions
Enter the object name, followed by a period, and the keyword cellipse. This should be followed by a list of the coefficient restrictions. Joint (multiple) coefficient restrictions should be separated by commas.
Options
ind=arg | Specifies whether and how to draw the individual coefficient intervals. The default is “ind=line” which plots the individual coefficient intervals as dashed lines. “ind=none” does not plot the individual intervals, while “ind=shade” plots the individual intervals as a shaded rectangle. |
size=number (default=0.95) | Set the size (level) of the confidence ellipse. You may specify more than one size by specifying a space separated list enclosed in double quotes. |
dist=arg | Select the distribution to use for the critical value associated with the ellipse size. The default depends on estimation object and method. If the parameter estimates are least-squares based, the 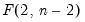 distribution is used; if the parameter estimates are likelihood based, the 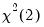 distribution will be employed. “dist=f” forces use of the F-distribution, while “dist=c” uses the 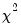 distribution. |
prompt | Force the dialog to appear from within a program. |
p | Print the graph. |
Examples
The two commands:
sys1.cellipse c(1), c(2), c(3)
sys1.cellipse c(1)=0, c(2)=0, c(3)=0
both display a graph showing the 0.95-confidence ellipse for C(1) and C(2), C(1) and C(3), and C(2) and C(3).
sys1.cellipse(dist=c,size="0.9 0.7 0.5") c(1), c(2)
displays multiple confidence ellipses (contours) for C(1) and C(2).
Cross-references
Clear the contents of the history attribute.
Removes the system’s history attribute, as shown in the label view of the system.
Syntax
system_name.clearhist
Examples
s1.clearhist
s1.label
The first line removes the history from the system S1, and the second line displays the label view of S1, including the now blank history field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Clear the contents of the remarks attribute.
Removes the system’s remarks attribute, as shown in the label view of the system.
Syntax
system_name.clearremarks
Examples
s1.clearremarks
s1.label
The first line removes the remarks from the system S1, and the second line displays the label view of S1, including the now blank remarks field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Coefficient covariance matrix.
Displays the covariances of the coefficient estimates for an estimated system.
Syntax
system_name.coefcov(options)
Options
p | Print the coefficient covariance matrix. |
Examples
sys1.coefcov
displays the coefficient covariance matrix for system SYS1 in a window. To store the coefficient covariance matrix as a sym object, use “@coefcov”:
sym eqcov = sys1.@coefcov
Cross-references
Creates a copy of the system.
Creates either a named or unnamed copy of the system.
Syntax
system_name.copy
system_name.copy dest_name
Examples
s1.copy
creates an unnamed copy of the system S1.
s1.copy s2
creates S2, a copy of the system S1.
Cross-references
Display graphs or tables of residual autocorrelations and cross-correlations.
Displays the auto and cross-correlation functions of the estimated system residals.
Syntax
system_name.correl(n, options)
You must specify the largest lag n to use in the computations. The default is to display a graphical view of the auto and cross-correlations.
Options
graph (default) | Display correlograms (graphs). |
bylag | Display table of results grouped by lag. |
bser | Display table of results grouped by series. |
factor=chol | Factorization by the inverse of the Cholesky factor of the residual covariance matrix (if estimated by ARCH). |
factor=cor | Factorization by the inverse square root of the residual correlation matrix (if estimated by ARCH; Doornik and Hansen, 1994). |
factor=cov | Factorization by the inverse square root of the residual covariance matrix (if estimated by ARCH; Urzua, 1997). |
name=arg | Save matrix of results. |
prompt | Force the dialog to appear from within a program. |
p | Print the correlograms. |
Examples
sys.correl(24)
Displays the correlograms of the SER1 series for up to 24 lags.
Cross-references
See
“Correlogram” and
“Cross Correlations and Correlograms” for related discussion of autocorrelation and cross-correlation functions, respectively. See also
“Residual Tests” for related testing in a VAR context.
Examine derivatives of the system equation specification.
Display information about the derivatives of the equation specification in tabular, graphical, or summary form.
The (default) summary form shows information about how the derivative of the equation specification was computed, and will display the analytic expression for the derivative, or a note indicating that the derivative was computed numerically. The tabular form shows a spreadsheet view of the derivatives of the regression specification with respect to each coefficient (for each observation). The graphical form of the view shows this information in a multiple line graph.
Syntax
system_name.derivs(options)
Options
g | Display multiple graph showing the derivatives of the equation specification with respect to the coefficients, evaluated at each observation. |
t | Display spreadsheet view of the values of the derivatives with respect to the coefficients evaluated at each observation. |
p | Print results. |
Note that the “g” and “t” options may not be used at the same time.
Examples
To show a table view of the derivatives:
sys1.derivs(t)
To display and print the summary view:
sys1.derivs(p)
Cross-references
See
“Derivative Computation” for details on the computation of derivatives.
See also
Equation::makederivs for additional routines for examining derivatives, and
System::grads, and
Equation::makegrads for corresponding routines for gradients.
Display table, graph, or spool output in the system object window.
Display the contents of a table, graph, or spool in the window of the system object.
Syntax
system_name.display object_name
Examples
system1.display tab1
Display the contents of the table TAB1 in the window of the object SYSTEM1.
Cross-references
Most often used in constructing an EViews Add-in. See
“Custom Object Output”.
Display name for system objects.
Attaches a display name to a system object which may be used to label output in place of the standard system object name.
Syntax
system_name.displayname display_name
Display names are case-sensitive, and may contain a variety of characters, such as spaces, that are not allowed in system object names.
Examples
hrs.displayname Hours Worked
hrs.label
The first line attaches a display name “Hours Worked” to the system object HRS, and the second line displays the label view of HRS, including its display name.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Displays a spreadsheet or graph view of the endogenous variables.
Syntax
system_name.endog(options)
Options
g | Multiple line graphs of the solved endogenous series. |
p | Print the table of solved endogenous series. |
Examples
sys1.endog(g,p)
prints the graphs of the solved endogenous series.
Cross-references
Displays the covariance matrix used in estimation.
The estimation covariance contains:
1. the identity matrix for OLS and TSLS
2. a diagonal matrix with equation variances used to compute WOLS and WTSLS
3. the residual covariance matrix used to compute SUR and 3SLS
4. the residual covariance matrix for unrestricted FIML; diagonal residual covariance matrix for diagonal FIML; user-specified covariance for user-covariance FIML
5. the long-run covariance of the moments used to compute the weighting matrix for GMM estimates
6. a matrix of missing values for ARCH
Syntax
system_name.estcov(options)
Options
p | Print the estimation covariance. |
Examples
sys1.estcov
displays the estimation covariance.
Cross-references
Estimation by full information maximum likelihood.
fiml estimates a system of equations by full information maximum likelihood (assuming a multivariate normal distribution).
Syntax
system_name.fiml(options)
Options
rcov = arg | Restricted residual covariance matrix in estimation: “diag” (non-zero diagonal and zero off-diagonal elements), “usercov” (fully specified user covariance matrix), “userfactor” user provides the matrix 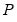 , such that 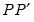 equals the fully specified covariance matrix). Note that system objects estimated using a restricted FIML estimator are not backward compatible with earlier versions of EViews, and will be dropped from the workfile if opened in a version prior to 9.5. |
rcovname= arg | Name of the matrix for determining the user specified residual covariance matrix. (Applicable when “rcov=usercov” or “rcov=userfactor”.) |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). “bfgs” is the default for new equations. |
optstep = arg | Step method: “marquardt” (Marquardt - default); “dogleg” (Dogleg); “linesearch” (Line search). (Applicable when “optmethod=bfgs”, “optmethod=newton” or “optmethod=opg”.) |
b | Use Berndt-Hall-Hall-Hausman (BHHH) as maximization algorithm. The default is Marquardt. (Applicable when “optmethod=legacy”.) |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “ (Applicable when non-legacy “optmethod=” with “cov=ordinary”.) |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
sys1.fiml
estimates SYS1 by FIML using the default settings.
sys1.fiml(rcov=diag)
estimates SYS1 by FIML with the off-diagonal residual covariances set to zero.
sys1.fiml(rcov=user, rcovname=mycov)
estimates a FIML model using MYCOV as the residual covariance matrix.
Cross-references
See
“System Estimation” for a discussion of systems in EViews.
Conditional variance/covariance of (G)ARCH estimation.
Displays the conditional variance, covariance or correlation of a system estimated by ARCH.
Syntax
system_name.garch(options) [arg1, arg2, ...]
The optional arguments following the keyword indicate which endogenous variable to include. If no argument is provided, all variables in the system will be included.
Options
cor | Display correlation. |
cov (default) | Display covariance. |
var | Display only variance. |
sd | Display only standard deviation. |
graph (default) | Display data in graph. |
mat | Display data in matrix format. |
list | Display data in list format. |
smpl=arg | Date to return conditional covariance value. |
pre | Include presample data (used with the mat option only). |
prompt | Force the dialog to appear from within a program. |
p | Print the graph |
Examples
sys1.garch(cor)
displays the conditional correlation graph of SYS1.
Cross-references
ARCH estimation is described in
“ARCH and GARCH Estimation”.
Estimation by generalized method of moments (GMM).
The system object must be specified with a list of instruments.
Syntax
system_name.gmm(options)
Options
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get the one-step weighting matrix. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
w | Use White’s diagonal weighting matrix (for cross section data). |
b=arg (default=“nw”) | Specify the bandwidth: “nw” (Newey-West fixed bandwidth based on the number of observations), number (user specified bandwidth), “v” (Newey-West automatic variable bandwidth selection), “a” (Andrews automatic selection). |
q | Use the quadratic kernel. Default is to use the Bartlett kernel. |
n | Prewhiten by a first order VAR before estimation. |
i | Iterate simultaneously over the weighting matrix and the coefficient vector. |
s | Iterate sequentially over the weighting matrix and coefficient vector. |
o (default) | Iterate only on the coefficient vector with one step of the weighting matrix. |
c | One step (iteration) of the coefficient vector following one step of the weighting matrix. |
e | TSLS estimates with GMM standard errors. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Note that some options are only available for a subset of specifications.
Examples
For system estimation, the command:
sys1.gmm(b=a, q, i)
estimates the system SYS1 by GMM with a quadratic kernel, Andrews automatic bandwidth selection, and iterates simultaneously over the weight and coefficient vectors until convergence.
Cross-references
See
“Additional Regression Tools” and
“System Estimation” for discussion of the various GMM estimation techniques.
Gradients of the objective function.
Displays the gradients of the objective function (where available) for an estimated system object.
The (default) summary form shows the value of the gradient vector at the estimated parameter values (if valid estimates exist) or at the current coefficient values. Evaluating the gradients at current coefficient values allows you to examine the behavior of the objective function at starting values. The tabular form shows a spreadsheet view of the gradients for each observation. The graphical form shows this information in a multiple line graph.
Syntax
system_name.grads(options)
Options
Examples
To show a summary view of the gradients:
sys1.grads
To print the table view:
sys1.grads(p)
Cross-references
Multivariate residual normality test.
Syntax
var_name.jbera(options)
You must specify a factorization method using the “factor=” option.
Options
factor=chol | Factorization by the inverse of the Cholesky factor of the residual covariance matrix. |
factor=cor | Factorization by the inverse square root of the residual correlation matrix (Doornik and Hansen, 1994). |
factor=cov | Factorization by the inverse square root of the residual covariance matrix (Urzua, 1997). |
name=arg | Save the test statistics in a named matrix object. See below for a description of the statistics contained in the stored matrix. |
prompt | Force the dialog to appear from within a program. |
p | Print the test results. |
The “
name=” option stores the following matrix. Let the VAR have
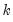
endogenous variables. Then the stored matrix will have dimension
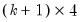
. The first
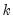
rows contain statistics for each orthogonal component, where the first column contains the third moments, the second column contains the
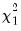
statistics for the third moments, the third column contains the fourth moments, and the fourth column holds the
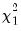
statistics for the fourth moments. The sum of the second and fourth columns are the Jarque-Bera statistics reported in the last output table.
The last row contains statistics for the joint test. The second and fourth column of the
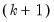
row is simply the sum of all the rows above in the corresponding column and are the
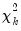
statistics for the joint skewness and kurtosis tests, respectively. These joint skewness and kurtosis statistics add up to the joint Jarque-Bera statistic reported in the output table, except for the “
factor=cov” option. When this option is set, the joint Jarque-Bera statistic includes all cross moments (in addition to the pure third and fourth moments). The overall Jarque-Bera statistic for this statistic is stored in the first column of the
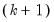
row (which will be a missing value for all other options).
Examples
sys01.jbera(factor=cor,name=jb)
carries out the residual multivariate normality test using the inverse square root of the residual correlation matrix as the factorization matrix and stores the results in a matrix named JB.
Cross-references
See
“Vector Autoregression (VAR) Models” for a discussion of the test in the context of VAR diagnostics.
Display or change the label view of the system object, including the last modified date and display name (if any).
As a procedure, label changes the fields in the system object label.
Syntax
system_name.label
system_name.label(options) [text]
Options
The first version of the command displays the label view of the system. The second version may be used to modify the label. Specify one of the following options along with optional text. If there is no text provided, the specified field will be cleared.
c | Clears all text fields in the label. |
d | Sets the description field to text. |
s | Sets the source field to text. |
u | Sets the units field to text. |
r | Appends text to the remarks field as an additional line. |
p | Print the label view. |
Examples
The following lines replace the remarks field of system S1 with “Data from CPS 1988 March File”:
s1.label(r)
s1.label(r) Data from CPS 1988 March File
To append additional remarks to S1, and then to print the label view:
s1.label(r) Log of hourly wage
s1.label(p)
To clear and then set the units field, use:
s1.label(u) Millions of bushels
Cross-references
See
“Labeling Objects” for a discussion of labels.
Estimation by linear or nonlinear least squares regression.
Syntax
system_name.ls(options)
Options
General options
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values for equations with AR or MA terms (see also
param). |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
sys1.ls(m=100)
estimates SYS1 using least squares, with the maximum number of iterations set at 100.
Cross-references
“Basic Regression Analysis” and
“Additional Regression Tools” discuss the various regression methods in greater depth.
See
“Special Expression Reference” for special terms that may be used in system
ls specifications.
Make a group out of the endogenous series.
Syntax
system_name.makeendog name
Following the keyword makeendog, you should provide a name for the group to hold the endogenous series. If you do not provide a name, EViews will create an untitled group.
Examples
sys1.makeendog grp_v1
creates a group named GRP_V1 that contains the endogenous series in SYS1.
Cross-references
Generate conditional variance series.
Saves the estimated conditional variance (from a system estimated using ARCH) as a named series. You may also save the conditional covariance or correlation.
Syntax
system_name.makegarch(options) [series1_name series2_name]
The optional series name arguments following the makegarch keyword indicate which endogenous variables to include. If no argument is given, all variables in the system will be included.
Options
cor | Generate conditional correlation. |
cov (default) | Generate conditional variance and covariance. |
var | Generate conditional variance. |
mat | Output as a matrix (default is to output as a series). |
name=arg | Base name or matrix name of the data to be saved. |
date=arg | Date to return conditional covariance value (used only with the mat option). |
pre | Include presample data (used only with the mat option). |
prompt | Force the dialog to appear from within a program. |
Examples
sys01.makegarch
creates conditional variances and conditional covariance series using the default names GARCH_01, GARCH_02, etc. for the conditional variance and GARCH_01_02, GARCH_01_03, etc. for the conditional covariance.
sys01.makegarch(mat, cor, date=12/11/2000, name=cov_mat)
creates a matrix named COV_MAT that contains the conditional correlation for the date 12/11/2000.
Cross-references
See
“ARCH and GARCH Estimation” for a discussion of GARCH models.
Create and save log likelihood contribution from system (ARCH estimation).
Syntax
system_name.makeloglike [ser1]
After the keyword, provide an optional name to save the log likelihood contribution. If you do not provide a name, EViews will name the series using the next available name of the form “LOGLIKE##”. (If LOGLIKE01 already exists, it will be named LOGLIKE02, and so on.)
Examples
sys1.makeloglike logl
creates a series of log likelihood contribution for the system and saves it in the series LOG1.
Make a model from a system of equations.
Syntax
system_name.makemodel(name) assign_statement
If you provide a name for the model in parentheses after the keyword, EViews will create the named model in the workfile. If you do not provide a name, EViews will open an untitled model window if the command is executed from the command line.
Examples
sys3.makemodel(sysmod) @prefix s_
makes a model named SYSMOD from the estimated system. SYSMOD includes an assignment statement “ASSIGN @PREFIX S_”. Use the command “show sysmod” or “sysmod.spec” to open the SYSMOD window.
Cross-references
See
“Models” for a discussion of specifying and solving models in EViews.
Create residual series.
Creates and saves residuals in the workfile from an estimated system object.
Syntax
system_name.makeresids(options) [residual_names]
Follow the system name with a period and the makeresids keyword, then provide a list of names to be given to the stored residuals. You should provide as many names as there are equations. If there are fewer names than equations, EViews creates the extra residual series with names RESID01, RESID02, and so on. If you do not provide any names, EViews will also name the residuals RESID01, RESID02, and so on.
Options
n=arg | Create group object to hold the residual series. |
chol | Standardized residuals factorized using the inverse of Cholesky factor of the (conditional) covariance matrix (for system ARCH). |
cor | Standardized residuals factorized using the inverse square root of the (conditional) correlation matrix (for system ARCH). |
cov | Standardized residuals factorized using the inverse square root of the (conditional) covariance matrix (for system ARCH). |
bn=arg | Base name used to generate the name of the residual series. |
prompt | Force the dialog to appear from within a program. |
Examples
sys1.makeresids res_sys1
creates a set of series containing the residuals from the system using RES_SYS1 to name the first equation residual, and RESID01, RESID02, etc., to name the remaining residuals.
Cross-references
Push updates to OLE linked objects in open applications.
Syntax
system_name.olepush
Cross-references
See
“Object Linking and Embedding (OLE)” for a discussion of using OLE with EViews.
Display estimation output.
output changes the default object view to display the estimation output (equivalent to using
System::results).
Syntax
system_name.output
Options
p | Print estimation output for estimation object |
Examples
The output keyword may be used to change the default view of an estimation object. Entering the command:
sys1.output
displays the estimation output for system SYS1.
Cross-references
Multivariate residual autocorrelation Portmanteau tests.
Syntax
system_name.qstats(h, options)
You must specify the highest order of lag h to test for serial correlation .
Options
maxlag=arg | Maximum lag in system specification (default=0). |
chol | Standardized residuals factorized using the inverse of Cholesky factor of the (conditional) covariance matrix (for system ARCH). |
cor | Standardized residuals factorized using the inverse square root of the (conditional) correlation matrix (for system ARCH). |
cov | Standardized residuals factorized using the inverse square root of the (conditional) covariance matrix (for system ARCH). |
prompt | Force the dialog to appear from within a program. |
p | Print the Portmanteau test results. |
Examples
show sys1.qstats(l0)
displays the portmanteau tests for lags up to 10.
Cross-references
See
“Diagnostic Views” for a discussion of the Portmanteau tests and other VAR diagnostics.
See
Var::arlm for a related multivariate residual serial correlation LM test.
Display text of specification for system objects.
Syntax
system_name.representation(options)
Options
p | Print the representation text. |
Examples
sys1.representations
displays the specifications of the equations in SYS1.
Residual correlation matrix.
Displays the correlations of the residuals from each equation in the system.
Syntax
system_name.residcor(options)
Options
p | Print the correlation matrix. |
Examples
sys1.residcor
displays the residual correlation matrix of SYS1.
Cross-references
Residual covariance matrix.
Displays the covariances of the residuals from each equation in the system.
Syntax
system_name.residcov(options)
Options
p | Print the covariance matrix. |
Examples
sys1.residcov
displays the residual covariance matrix of SYS1.
Cross-references
Display residuals.
resids displays multiple graphs or a spreadsheet of the residuals. Each graph will contain the residuals for each equation in the system.
Syntax
system_name.resids(options)
Options
sheet | Display residuals in spreadsheet. |
p | Print the table/graph. |
Examples
sys1.resids
displays a graph of the residual series in system SYS1.
Cross-references
Displays the results view of an estimated system.
Syntax
system_name.results(options)
Options
Examples
sys1.results(p)
displays and prints the results of SYS1.
Set the object attribute.
Syntax
system_name.setattr(attr) attr_value
Sets the attribute attr to attr_value. Note that quoting the arguments may be required. Once added to an object, the attribute may be extracted using the @attr data member.
Examples
a.setattr(revised) never
String s = a.@attr("revised")
sets the “revised” attribute in the object A to the string “never”, and extracts the attribute into the string object S.
Cross-references
Display the text specification view for system objects.
Syntax
system_name.spec(options)
Options
p | Print the specification text. |
Examples
sys1.spec
displays the specification of the system object SYS1.
Cross-references
Estimate a system object using seemingly unrelated regression (SUR).
Note that the EViews procedure is more general than textbook versions of SUR since the system of equations may contain cross-equation restrictions on parameters.
Syntax
system_name.sur(options)
Options
i | Iterate on the weighting matrix and coefficient vector simultaneously. |
s | Iterate on the weighting matrix and coefficient vector sequentially. |
o (default) | Iterate only on the coefficient vector with one step of the weighting matrix. |
c | One step iteration on the coefficient vector after one step of the weighting matrix. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion.The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get one-step weighting matrix. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
sys1.sur(i)
estimates SYS1 by SUR, iterating simultaneously on the weighting matrix and coefficient vector.
nlsys.sur(showopts,m=500)
estimates NLSYS by SUR with up to 500 iterations. The “showopts” option displays the starting values.
Cross-references
See
“System Estimation” for a discussion of system estimation.
Declare system of equations.
Syntax
system system_name
Follow the system keyword by a name for the system. If you do not provide a name, EViews will open an untitled system object (if in interactive mode).
Examples
system mysys
creates a system named MYSYS.
Cross-references
“System Estimation” provides a full discussion of system objects.
See
System::append for adding specification lines to an existing system.
Two-stage least squares.
Syntax
system_name.tsls(options)
There must be at least as many instrumental variables as there are independent variables. All exogenous variables included in the regressor list should also be included in the instrument list. A constant is included in the list of instrumental variables even if not explicitly specified.
Options
General options
m=integer | Set maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
i | Iterate on the weighting matrix and coefficient vector simultaneously. |
s | Iterate on the weighting matrix and coefficient vector sequentially. |
o (default) | Iterate only on the coefficient vector with one step of the weighting matrix. |
c | One step iteration of the coefficient vector after one step of the weighting matrix. |
l=number | Set maximum number of iterations on the first-stage iteration to get one-step weighting matrix. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
sys1.tsls
estimates the system object using TSLS.
Cross-references
See
“Two-Stage Least Squares” for details on two-stage least squares estimation in systems.
See also
System::ls. For estimation of weighted TSLS in systems, see
System::wtsls.
Update coefficient object values from system object.
Copies coefficients from the system into the appropriate coefficient vector or vectors.
Syntax
system_name.updatecoefs
Follow the name of the system object by a period and the keyword updatecoefs.
Examples
SYS1.updatecoefs
places the coefficients from SYS1 in the coefficient vectors used in the system.
Cross-references
Wald coefficient restriction test.
The wald view carries out a Wald test of coefficient restrictions for a system object.
Syntax
system_name.wald restrictions
Enter the system name, followed by a period, and the keyword. You must provide a list of the coefficient restrictions, with joint (multiple) coefficient restrictions separated by commas.
Options
Examples
sys1.wald c(2)=c(3)*c(4)
tests the non-linear restriction that the second coefficient is equal to the product of the third and fourth coefficients in SYS1.
Cross-references
Estimates a system of equations using weighted least squares.
Syntax
system_name.wls(options)
Options
i | Iterate simultaneously over the weighting matrix and coefficient vector. |
s | Iterate sequentially over the computation of the weighting matrix and the estimation of the coefficient vector. |
o (default) | Iterate the estimate of the coefficient vector to convergence following one-iteration of the weighting matrix. |
c | One step (iteration) of the coefficient vector estimates following one iteration of the weighting matrix. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage coefficient estimation to get one-step weighting matrix. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print the estimation results. |
Examples
sys1.wls
estimates the system of equations in SYS1 by weighted least squares.
Cross-references
See
“System Estimation” for a discussion of system estimation.
See also the available options for weighted least squares in
System::ls.
Perform weighted two-stage least squares estimation of a system of equations.
Syntax
system_name.wtls(options)
Options
i | Iterate simultaneously over the weighting matrix and coefficient vector. |
s | Iterate sequentially over the computation of the weighting matrix and the estimation of the coefficient vector. |
o (default) | Iterate the coefficient vector to convergence following one-iteration of the weighting matrix. |
c | One step (iteration) of the coefficient vector following one iteration of the weighting matrix. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get the one-step weighting matrix. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
sys1.wtsls
estimates the system of equations in SYS1 by weighted two-stage least squares.
Cross-references
See also
System::tsls for both unweighted and weighted single equation 2SLS.
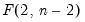 distribution is used; if the parameter estimates are likelihood based, the
distribution is used; if the parameter estimates are likelihood based, the 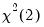 distribution will be employed. “dist=f” forces use of the F-distribution, while “dist=c” uses the
distribution will be employed. “dist=f” forces use of the F-distribution, while “dist=c” uses the 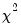 distribution.
distribution.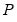 , such that
, such that 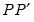 equals the fully specified covariance matrix).
equals the fully specified covariance matrix).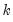 endogenous variables. Then the stored matrix will have dimension
endogenous variables. Then the stored matrix will have dimension 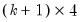 . The first
. The first 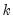 rows contain statistics for each orthogonal component, where the first column contains the third moments, the second column contains the
rows contain statistics for each orthogonal component, where the first column contains the third moments, the second column contains the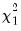 statistics for the third moments, the third column contains the fourth moments, and the fourth column holds the
statistics for the third moments, the third column contains the fourth moments, and the fourth column holds the 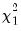 statistics for the fourth moments. The sum of the second and fourth columns are the Jarque-Bera statistics reported in the last output table.
statistics for the fourth moments. The sum of the second and fourth columns are the Jarque-Bera statistics reported in the last output table. 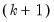 row is simply the sum of all the rows above in the corresponding column and are the
row is simply the sum of all the rows above in the corresponding column and are the 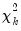 statistics for the joint skewness and kurtosis tests, respectively. These joint skewness and kurtosis statistics add up to the joint Jarque-Bera statistic reported in the output table, except for the “
statistics for the joint skewness and kurtosis tests, respectively. These joint skewness and kurtosis statistics add up to the joint Jarque-Bera statistic reported in the output table, except for the “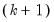 row (which will be a missing value for all other options).
row (which will be a missing value for all other options).