Equation
Equation object. Equations are used for single equation estimation, testing, and forecasting.
Equation Declaration
To declare an equation object, enter the keyword equation, followed by a name:
equation ecoefq01
and an optional specification:
equation r4cst.ls r c r(-1) div
equation wcd.ls q=c(1)*n^c(2)*k^c(3)
Equation Methods
arch autoregressive conditional heteroskedasticity (ARCH and GARCH).
ardl autoregressive distributed lag models.
binary binary dependent variable models (includes probit, logit, gompit) models.
breakls least squares with breakpoints and breakpoint determination.
censored censored and truncated regression (includes tobit) models.
cointreg cointegrating regression using FMOLS, CCR, or DOLS, or panel FMOLS or DOLS.
count count data modeling (includes poisson, negative binomial and quasi-maximum likelihood count models).
did estimate a panel equation using the difference-in-difference estimator.
enet elastic net regression (including Lasso and ridge regression).
funcoef functional coefficients regression.
glm estimate a Generalized Linear Model (GLM).
gmm estimate an equation using generalized method of moments (GMM).
heckit estimate a selection equation using the Heckman ML or 2-step method.
liml estimate an equation using Limited Information Maximum Likelihood and K-class estimation.
logit logit (binary) estimation.
ls estimation using least squares or nonlinear least squares.
midas Mixed Data Sampling (MIDAS) regression.
ordered ordinal dependent variable models (includes ordered probit, ordered logit, and ordered extreme value models).
probit probit (binary) estimation.
qreg estimate an equation using quantile regression.
robustls robust regression (M-estimation, S-estimation and MM-estimation).
switchreg exogenous and Markov switching regression.
threshold threshold least squares, including threshold autoregression.
tsls estimate an equation using two-stage least squares regression.
varsel equation estimation using least squares with variable selection (uni-directional, stepwise, swapwise, combinatorial, Auto-GETS, Lasso).
Equation Views
abtest test for serial correlation in a panel GMM equation using the Arellano-Bond test.
archtest LM test for the presence of ARCH in the residuals.
arma Examine ARMA structure of estimated equation.
auto Breusch-Godfrey serial correlation Lagrange Multiplier (LM) test.
boundstest perform the Pesaran, Shin and Smith (2001) bounds test of long-run relationships from an ARDL estimated equation.
breakspec display the breakpoint specification for an equation estimated by least squares with breakpoints.
breaktest perform breakpoint test for TSLS and GMM equations.
cdtest test for the presence of cross-sectional dependence of errors in panel equations.
cellipse confidence ellipses for coefficient restrictions.
chow Chow breakpoint and forecast tests for structural change.
cinterval confidence interval for coefficients.
coefcov coefficient covariance matrix.
coeflabel display coefficients associated with variables in the equation.
coefmatrix display matrix of lambda and coefficients for elastic net, ridge, and Lasso models.
coefpath display graphs of the paths of the coefficients plotted against lambda, fit measures, and estimation values
in elastic net, ridge, Lasso, and variable selection using Lasso models.
coint test for cointegration between series in an equation estimated using cointegrating regression.
cointgraph view a graph of the estimated cointegrating relation form of an ARDL estimated equation.
cointrel display information about the cointegrating relation specification and the coefficients in ARDL estimated equation.
cointrep view the estimated cointegration form and the long-run coefficients table of an ARDL estimated equation.
correl correlogram of the residuals.
correlsq correlogram of the squared residuals.
cvardecomp coefficient covariance decomposition table.
cvgraph display a graph of the cross-validation objective against the lambda path
for elastic net, ridge, Lasso, and variable selection using Lasso models.
depfreq display frequency and cumulative frequency table for the dependent variable.
derivs derivatives of the equation specification.
didcs compute Callaway-Sant’Anna decomposition for difference-in-difference estimation.
didgbdecomp perform Goodman-Bacon decomposition for difference-in-difference estimation.
didtrends show difference-in-difference trends summary in graphical or tabular form.
display display table, graph, or spool in object window.
dynmult compute dynamic multipliers for long-run regressors in ARDL equations.
ecresults display the conditional error correction (CEC) and error correction (EC) regression results.
effects display table of estimated fixed and/or random effects.
endogtest perform the regressor endogeneity test.
facbreak factor breakpoint test for stability.
resoutliers detect outliers in the residuals or regressors of the equation.
fixedtest test significance of estimates of fixed effects for panel estimators.
funbias functional coefficients equation bias results.
funbw functional coefficients equation bandwidth results.
funci functional coefficients equation coefficient confidence intervals.
funcov functional coefficients covariance results.
funtest perform functional coefficients hypothesis and stability tests.
garch conditional standard deviation graph (only for equations estimated using ARCH).
grads examine the gradients of the objective function.
hettest test for heteroskedasticity.
hist histogram and descriptive statistics of the residuals.
icgraph display a graph of the selection criteria for the top 20 models observed as part of model selection during estimation.
ictable display a table of the log-likelihood and selection criteria for the top 20 models observed as part of model selection during estimation.
infbetas scaled difference in estimated betas for influence statistics.
instsum show a summary of the equation instruments.
label label information for the equation.
lambdacoefs display the spreadsheet of the matrix of coefficient values along the lambda path in elastic net, ridge, Lasso, and variable selection using Lasso models.
lambdaest display the table showing various values associated with estimation along the lambda path in elastic net, ridge, Lasso, and variable selection using Lasso models.
lambdafit display the table showing various fit statistics associated with estimates along the lambda path in elastic net, ridge, Lasso, and variable selection using Lasso models.
lambdapath display graphs of lambda against various fit and estimation measures in elastic net, ridge, Lasso, and variable selection using Lasso models.
means descriptive statistics by category of the dependent variable (only for binary, ordered, censored and count equations).
modselgraph display a graph of the selection criteria for the top 20 models for elastic net, ridge, Lasso, and variable selection using Lasso models.
modseltable display a table of the selection criteria and measures associated with the estimation and model selection of elastic net, ridge, Lasso, and variable selection using Lasso models.
multibreak perform multiple breakpoint testing for an equation specified by list and estimated by least squares.
newsimpact display a news-impact graph of equations estimated using GARCH.
nyblom perform the Nyblom test of parameter stability or structural change in equations estimated using GARCH.
orthogtest perform the instrument orthogonality test.
outliers display the outliers summary view for an equation estimated via least squares with automatic outlier indicator saturation.
output table of estimation results.
pmghausmantest displays a spool object with the results of the Hausman test for similarity against mean-group and dynamic fixed effects estimators in PMG estimation.
predict prediction (fit) evaluation table (only for binary and ordered equations).
qrcrprocess displays a spool object producing a quantile process of the cointegrating relation.
qrecprocess displays a spool object producing a quantile process for each of the conditional error correction and error correction coefficients.
qrprocess display table or graph of quantile process estimates.
qrslope test of equality of slope coefficients across multiple quantile regression estimates.
qrsymm test of coefficients using symmetric quantiles.
ranhaus Hausman test for correlation between random effects and regressors.
rcomptest tests for the presence of cross-sectional or time random components in a panel equation. estimated using pooled least squares.
reset Ramsey’s RESET test for functional form.
resids display, in tabular form, the actual and fitted values for the dependent variable, along with the residuals.
resoutliers Detect outliers in the residuals or regressors of the equation.
results table of estimation results.
rgmprobs display the regime probabilities in a switching regression equation.
rls recursive residuals least squares (only for non-panel equations estimated by ordinary least squares, without ARMA terms).
signbias perform the Sign bias test (Engle and Ng, 1993) of misspecification in equations estimated using GARCH.
similarity compute Hausman tests for Pooled mean group ARDL equations.
srcoefs displays a spool object with the results of error-correction regressions for each cross-section in PMG estimation.
strconstant tests for constancy of the base specification coefficients against a smoothly varying alternative in a smooth threshold regression.
strlinear compute tests for linearity of the base specification against the smooth threshold alternative in a smooth threshold regression.
strnonlin compute various tests for additional additive or encapsulated nonlinearity in a smooth threshold regression.
strwgts compute and display the transition weights in a smooth threshold regression.
symmtest compute symmetry test for nonlinear distributed lag variables in nonlinear ARDL models.
testadd likelihood ratio test for adding variables to equation.
testdrop likelihood ratio test for dropping variables from equation.
testfit performs Hosmer and Lemeshow and Andrews goodness-of-fit tests (only for equations estimated using binary).
transprobs display the state transition probabilities in a switching regression equation.
ubreak Andrews-Quandt test for unknown breakpoint.
varinf display Variance Inflation Factors (VIFs).
wald Wald test for coefficient restrictions.
weakinst display the weak instruments summary.
white White test for heteroskedasticity.
Equation Procs
clearhist clear the contents of the history attribute.
copy creates a copy of the equation.
didmakeeq create an equation object with the underlying fixed-effects estimation of a difference-in-difference equation.
makecoint Create a series containing the estimated cointegrating relationship from an ARDL estimated equation.
makederivs make group containing derivatives of the equation specification.
makefunobj save coefficients, residuals, bias, variance, and confidence intervals for functional coefficients equations.
makegarch create conditional variance series (only for ARCH equations).
makegrads make group containing gradients of the objective function.
makelimits create vector of estimated limit points (only for ordered models).
makemodel create model from estimated equation.
makeregs make group containing the regressors.
makergmprobs save the regime probabilities in a switching regression equation.
makeresids make series containing residuals from equation.
makestrwgts save the smooth transition weights in a smooth threshold regression.
maketransprobs save the state transition probabilities in a switching regression equation.
olepush push updates to OLE linked objects in open applications.
setattr set the value of an object attribute.
setpilotbw compute and set the value of the local pilot bandwidth (for functional coefficients equations).
updatecoefs update coefficient vector(s) from equation.
Equation Data Members
Scalar Values
@aic Akaike information criterion.
@bylist returns 1 or 0 depending on whether the equation was estimated by list.
@coefcov(i,j) covariance of coefficient estimates i and j.
@coefs(i) i-th coefficient value.
@deviance deviance (for Generalized Linear Models).
@deviancestat deviance statistic: deviance divided by degrees-of-freedom (for Generalized Linear Models).
@df degrees-of-freedom for equation.
@dispersion estimate of dispersion (for Generalized Linear Models).
@dw Durbin-Watson statistic.
@f F-statistic.
@finalbw returns the final bandwidth used in functional coefficient estimation.
@fixeddisp indicator for whether the dispersion is a fixed value (for Generalized Linear Models).
@fprob probability value of the F-statistic.
@hacbw bandwidth for HAC estimation of GMM weighting matrix or long-run covariance in cointegrating regression (if applicable).
@hq Hannan-Quinn information criterion.
@instrank rank of instruments (if applicable).
@jstat J-statistic — value of the GMM objective function (for GMM and TSLS).
@jprob probability value of the J-statistic.
@lambdamin minimum lambda value from ENET cross-validation.
@limlk estimate of LIML
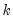
(if applicable).
@logl value of the log likelihood function.
@lrprob probability value of likelihood ratio statistic (if applicable).
@lrstat likelihood ratio statistic (if applicable).
@lrvar long-run variance estimate for cointegrating regression (if applicable).
@meandep mean of the dependent variable.
@nbreaks number of breaks in breakpoint least squares and thresholds in threshold regression.
@ncases number of cases.
@nclusters number of clusters used in computing cluster robust covariances.
@ncoef number of estimated coefficients.
@ncross number of cross-sections used in estimation (equal to 1 for non-panel workfiles).
@npers number of workfile periods used in estimation (same as @regobs for non-panel workfiles).
@nregimes number of regimes in a switching and breakpoint regression.
@nthresholds number of thresholds in threshold regression.
@ntreatment difference-in-difference number of cross sections receiving treatment.
@objective quasi-likelihood objective function (if applicable).
@pearsonssr Pearson sum-of-squared residuals (for Generalized Linear Models).
@pearsonstat Pearson statistic: Pearson SSR divided by degrees-of-freedom (for Generalized Linear Models).
@pilotbw returns the pilot bandwidth used in functional coefficient estimation.
@qlrprob probability value of quasi-likelihood ratio statistic (if applicable).
@qlrstat quasi-likelihood ratio statistic (if applicable).
@quantdep quantile of dependent variable (for quantile regression).
@r2 R-squared statistic.
@rbar2 adjusted R-squared statistic.
@rdeviance restricted (constant only) deviance (for Generalized Linear Models).
@regobs number of observations in regression.
@rlogl restricted (constant only) log-likelihood (if applicable).
@rmse root MSE.
@rn2 Rn-squared statistic.
@robf robust F-statistic (Wald-test form).
@robfprob robust F-statistic (Wald-test form) p-value.
@robjective restricted (constant only) quasi-likelihood objective function (if applicable).
@rw2 Rw-squared.
@schwarz Schwarz information criterion.
@sddep standard deviation of the dependent variable.
@se standard error of the regression.
@sparsity estimate of sparsity (for quantile regression).
@ssr sum of squared residuals.
@ssr2 second-stage SSR.
@stderrs(i) standard error for coefficient i.
@thresholds number of thresholds (for threshold regression).
@tstats(i) t-statistic or z-statistic value for coefficient i.
@wmeandep weighted mean of dependent variable (if applicable).
@wgtscale scaling factor for weights (if applicable).
c(i) i-th element of default coefficient vector for equation (if applicable).
Vectors and Matrices
@ardlceccoefs returns a vector of coefficient estimates from the conditional error-correction (CEC) regression in univariate (N)ARDL estimation.
@ardleccoefs returns a vector of coefficient estimates from the traditional error-correction (EC) regression in univariate (N)ARDL estimation.
@ardlcoint returns a coef containing coefficients from the cointegrating relationship form of an ARDL estimation.
@ardllrcoefs returns a coef containing coefficients from the long run relationship form of a non-panel ARDL estimation.
@ardlsrcoefs returns a matrix where each row corresponds to an individual cross-section’s short-run coefficients. Only applicable for PMG/ARDL estimation.
@ardlsrses returns a matrix where each row corresponds to an individual cross-section’s short-run coefficient standard errors. Only applicable for PMG/ARDL estimation.
@coefcov covariance matrix for coefficient estimates.
@coefs coefficient vector.
@contempcov symmetric matrix containing the contemporaneous covariance
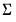
for cointegrating regression residuals estimated with CCR.
@cvconverge Elastic net path cross-validation convergence test values (lambda values in rows; lambda in first column, training-test sample results in remaining columns).
@cvisvalid Elastic net path cross-validation valid results indicators (lambda values in rows; lambda in first column, training-test sample results in remaining columns).
@cviters Elastic net path cross-validation iterations (lambda values in rows; lambda in first column, training-test sample results in columns).
@cvobjective Elastic net path cross-validation objective values (lambda values in rows; lambda in first column, training-test sample results in remaining columns).
@effects vector of fixed and random effects estimates (if applicable).
@fcgrid returns a vector of unique grid values over which functional coefficients are evaluated in functional coefficient estimation.
@initprobs matrix containing initial probabilities for switching regression equations.
@instwgt symmetric matrix containing the final sample instrument weighting matrix used during GMM or TSLS estimation (
e.g.,
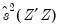
for 2SLS and
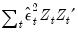
for White weighting).
@lambdacoefs Elastic net lambda path coefficients matrix (lambda values in rows; variables in columns). Full set of variables including those with zero coefficients along the path.
@lambdaest Elastic net lambda path estimation measures matrix (lambda values in rows; columns contain the lambda values, number of non-zero coefficients, estimation objective, sums-of-squares portion of the objective,
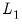
portion of the objective,
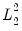
portion of the objective).
@lambdafit Elastic net lambda path fit measures matrix (lambda values in rows; columns contain the lambda values, number of non-zero coefficients, R-squared, adjusted R-squared, and sums-of-squared residuals).
@lambdapath Elastic net lambda path vector.
@lambda2cov symmetric matrix
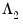
containing the long run covariances of
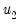
with
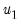
and
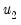
for cointegrating regression equations estimated with CCR and FMOLS.
@lrcov symmetric matrix containing the long-run covariance
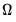
for cointegrating regression equations estimated with CCR and FMOLS.
@modselresults Elastic net path model selection summary (lambda values in rows; lambda in first column, followed by model selection objective, number of non-zero coefficients, and the fit statistics (sum-of-squared residuals, mean-square error, R-squared, and adjusted R-squared) associated with the estimated model.
@pmgcxcoefs returns a matrix of coefficient estimates from the error-correction regressions for each cross-section in PMG estimation. Each column corresponds to a single cross-section and each column corresponds to a coefficient estimate from the traditional error-correction regression, in order of appearance.
@pmgcxses returns a matrix of coefficient standard error estimates from the error-correction regressions for each cross-section in PMG estimation. Each column corresponds to a single cross-section and each column corresponds to a coefficient standard error estimate from the traditional error-correction regression, in order of appearance.
@pmglrcoefs returns a vector of long-run (pooled) coefficient estimates in PMG estimation.
@pmgsrcoefs returns a vector of short-run (mean-group) coefficient estimates in PMG estimation.
@pvals vector containing the coefficient probability values.
@stderrs vector of standard errors for coefficients.
@thresholds vector of threshold values for threshold estimation.
@tstats vector of t-statistic or z-statistic values for coefficients.
String Values
@ardlcointsubst returns string representation of the cointegration form of an ARDL equation with substituted coefficients.
@attr("arg") string containing the value of the arg attribute, where the argument is specified as a quoted string.
@breaks string representation of the breakpoints in breakpoint least squares or thresholds in threshold regression.
@coeflabels coefficient labels used in regression output table.
@coeflist returns a string containing a space delimited list of the coefficients used in estimation (e.g., “C(1) C(2) C(3)”). This function always returns the list of actual coefficients used, irrespective of whether the original equation was specified by list or by expression.
@command full command line form of the estimation command. Note this is a combination of @method, @options and @spec.
@depends string containing a list of the series in the current workfile on which this equation depends.
@description string containing the Equation object’s description (if available).
@detailedtype returns a string with the object type: “EQUATION”.
@displayname returns the equation’s display name. If the equation has no display name set, the name is returned.
@esteq returns string representation of the estimation equation.
@extralist space delimited list of the equation's extra regressors. For equation's estimated by ARCH, @extralist contains the variance equation terms. For equations estimated by CENSORED, this contains the error distribution terms. For all other equation methods it returns an empty string.
@instlist space delimited list of the equation instruments (if applicable).
@method command line form of estimation method (“ARCH”, “LS”, etc....).
@name returns the name of the Equation.
@options command line form of estimation options.
@remarks string containing the equation object’s remarks (if available).
@smpl description of the sample specified for estimation.
@spec original equation specification. Note this will be different from @varlist if the equation specification contains groups, or is specified by expression.
@subst returns string representation of the equation with substituted coefficients.
@type returns a string with the object type: “EQUATION”.
@updatetime returns a string representation of the time and date at which the equation was last updated.
@varlist space delimited list of the equation’s dependent variable and regressors if the equation was specified by list, or the equation’s underlying variables (both dependent and independent) if the equation was specified by expression.
@varselkept space delimited list of variables kept by model selection.
@varselrejected space delimited list of the variables dropped by model selection.
Equation Examples
To apply an estimation method (proc) to an existing equation object:
equation ifunc
ifunc.ls r c r(-1) div
To declare and estimate an equation in one step, combine the two commands:
equation value.tsls log(p) c d(x) @ x(-1) x(-2)
equation drive.logit ifdr c owncar dist income
equation countmod.count patents c rdd
To estimate equations by list, using ordinary and two-stage least squares:
equation ordinary.ls log(p) c d(x)
equation twostage.tsls log(p) c d(x) @ x(-1) x(-2)
You can create and use other coefficient vectors:
coef(10) a
coef(10) b
equation eq01.ls y=c(10)+b(5)*y(-1)+a(7)*inc
The fitted values from EQ01 may be saved using,
series fit = eq01.@coefs(1) + eq01.@coefs(2)*y(‑1) + eq01.@coefs(3)*inc
or by issuing the command:
eq01.fit fitted_vals
To perform a Wald test:
eq01.wald a(7)=exp(b(5))
You can save the t-statistics and covariance matrix for your parameter estimates:
vector eqstats=eq01.@tstats
matrix eqcov=eq01.@coefcov
Test for serial correlation in a panel GMM equation using the Arellano-Bond test.
Tests for first and second order autocorrelation amongst the residuals of an equation estimated by GMM with first differences in a panel workfile. If the underlying errors are i.i.d, we would expect the first differences to be negatively first order serially correlated, and not display second order correlation.
Syntax
eq_name.abtest(options)
Options
p | Print output from the test. |
Examples
equation eq1.gmm(cx=fd, per=f, gmm=perwhite, iter=oneb, levelper) n n(-1) n(-2) w w(-1) k ys ys(-1) @ @dyn(n,-2) w w(-1) k ys ys(-1)
eq1.abtest
estimates an equation using GMM with first difference fixed effects, and then tests for first and second order autocorrelation.
Cross-references
Estimate generalized autoregressive conditional heteroskedasticity (GARCH) models.
Syntax
eq_name.arch(p,q,options) y [x1 x2 x3] [@ p1 p2 [@ t1 t2]]
eq_name.arch(p,q,options) y=expression [@ p1 p2 [@ t1 t2]]
The first two options specify the order of the GARCH model:
• The arch estimation method specifies a GARCH(p, q) model with p ARCH terms and q GARCH terms. Note the order of the arguments in which the ARCH and GARCH terms are entered.
The maximum value for
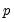
or
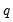
is 9; values above will be set to 9. The minimum value for
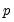
is 1. The minimum value for
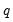
is 0. If either
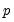
or
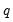
is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default.
• For CGARCH, FIEGARCH and MIDAS-GARCH models, EViews only estimates (1,1) models. For these specifications,
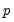
and

options should not be specified, and if provided, will be ignored.
After the “ARCH” keyword and options, specify the dependent variable followed by a list of regressors in the mean equation.
• By default, only the intercept is included in the conditional variance equation. If you wish to specify variance regressors, list them after the mean equation using an “@”-sign to separate the mean from the variance equation.
• When estimating component ARCH models, you may specify exogenous variance regressors for both the permanent and transitory components. After the mean equation regressors, first list the regressors for the permanent component, followed by an “@”-sign, then the regressors for the transitory component. A constant term is always included as a permanent component regressor.
• For MIDAS-GARCH models, the low-frequency permanent component regressor are entered after the mean equation regressors and an “@”-sign. The regressor should be specified as pagename\seriesname.
Options
Type Options
The default is to estimate a standard GARCH model. You may specify one of the followings keywords to estimate a different model:
egarch | Exponential GARCH. |
parch[=arg] | Power ARCH. If the optional arg is provided, the power parameter will be set to that value, otherwise the power parameter will be estimated. |
cgarch | Component (permanent and transitory) ARCH. |
figarch | Fractional GARCH (FIGARCH). |
fiegarch | Fractional Exponential GARCH (FIEGARCH(1,1)). |
midas | MIDAS GARCH(1,1) |
General Options
thrsh | For Component GARCH models, include a threshold term. |
thrsh=integer (default=0) | Number of threshold terms for GARCH models. The maximum number of terms allowed is 9. |
vt | Variance target of the constant term for GARCH models. (May not be used with integrated specifications.) |
integrated | Restrict GARCH model to be integrated, i.e. IGARCH. (May not be used with variance targeting.) |
asy=integer (default=1) | Number of asymmetric terms in Power ARCH or EGARCH models. The maximum number of terms allowed is 9. |
trunclag=integer (default=1000) | Number of terms in the expansion approximation for FIGARCH and FIEGARCH models. |
archm=arg | ARCH-M (ARCH in mean) specification with the conditional standard deviation (“archm=sd”), the conditional variance (“archm=var”), or the log of the conditional variance (“archm= log”) entered as a regressor in the mean equation. |
tdist [=number] | Estimate the model assuming that the residuals follow a conditional Student’s t-distribution (the default is the conditional normal distribution). Providing the optional number greater than two will fix the degrees of freedom to that value. If the argument is not provided, the degrees of freedom will be estimated. |
ged [=number] | Estimate the model assuming that the residuals follow a conditional GED (the default is the conditional normal distribution). Providing a positive value for the optional argument will fix the GED parameter. If the argument is not provided, the parameter will be estimated. |
z | Turn of backcasting for both initial MA innovations and initial variances. |
backcast=n | Backcast weight to calculate value used as the presample conditional variance. Weight needs to be greater than 0 and less than or equal to 1; the default value is 0.7. Note that a weight of 1 is equivalent to no backcasting, i.e. using the unconditional residual variance as the presample conditional variance. |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). “bfgs” is the default for new equations. |
optstep = arg | Step method: “marquardt” (Marquardt - default); “dogleg” (Dogleg); “linesearch” (Line search). (Applicable when “optmethod=bfgs”, “optmethod=newton” or “optmethod=opg”.) |
b | Use Berndt-Hall-Hall-Hausman (BHHH) as maximization algorithm. The default is Marquardt. (Applicable when “optmethod=legacy”.) |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “bollerslev” (Bollerslev-Wooldridge method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “ (Applicable when non-legacy “optmethod=” with “cov=ordinary”.) |
h | Bollerslev-Wooldridge robust quasi-maximum likelihood (QML) covariance/standard errors. (Applicable for “optmethod=legacy” when estimating assuming normal errors.) |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of preliminary LS estimates (out of range values are set to “s=1”). |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. Available only for legacy estimation (“optmeth=legacy”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
MIDAS Options
lag=arg | Specify the number of lags of the low frequency regressor to include. Default value is 32. |
beta=arg | Beta function restriction: none (“none”), trend coefficient equals 1 (“trend”), endpoints coefficient equals 0 (“end-point”), both trend and endpoints restriction (“both”). For use when “midwgt=beta”. The default is “beta=none”. |
thrsh | Include a threshold term. |
optmethod=arg | Optimization method for nonlinear estimation: “bfgs” (BFGS); “newton” Newton-Raphson), “opg”, “bhhh” (OPG or BHHH), or “hybrid” (initial BHHH followed by BFGS). Hybrid is the default method. |
optstep=arg | Step method for nonlinear estimation: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method for nonlinear models: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich). |
covinfo=arg | Information matrix method for nonlinear models: “opg” (OPG); “hessian” (observed Hessian). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values in nonlinear estimation. If the “s=number” or “s” options are not used, EViews will use random starting values. |
s=number | Determine starting values for nonlinear estimation. Specify a number between zero and oSpecify the number of lags of the low frequency regressor to include. Default value is 32.ne representing the fraction of preliminary EViews chosen values. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. If the “s=number” or “s” options are not used, EViews will use random starting values. |
seed=positive integer from 0 to 2,147,483,647 | Seed the random number generator used in random starting values. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
showopts/-showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector; the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Saved results
Most of the results saved for the
ls estimation method are also available after ARCH estimation; see
Equation::ls for details.
Examples
equation arc1.arch(4, 0, m=1000, cov=bollerslev) sp500 c
estimates an ARCH(4) model with a mean equation consisting of the series SP500 regressed on a constant. The procedure will perform up to 1000 iterations, and will report Bollerslev-Wooldridge robust QML standard errors upon completion.
The commands:
c = 0.1
equation arc1.arch(thrsh=1, s, mean=var) @pch(nys) c ar(1)
estimate a TARCH(1, 1)-in-mean specification with the mean equation relating the percent change of NYS to a constant, an AR term of order 1, and a conditional variance (GARCH) term. The first line sets the default coefficient vector to 0.1, and the “s” option uses these values as coefficient starting values.
The command:
equation arc1.arch(1, 2, asy=0, parch=1.5, ged=1.2) dlog(ibm)=c(1)+c(2)* dlog(sp500) @ r
estimates a symmetric Power ARCH(2, 1) (autoregressive GARCH of order 2, and moving average ARCH of order 1) model with GED errors. The power of model is fixed at 1.5 and the GED parameter is fixed at 1.2. The mean equation consists of the first log difference of IBM regressed on a constant and the first log difference of SP500. The conditional variance equation includes an exogenous regressor R.
Following estimation, we may save the estimated conditional variance as a series named GARCH1.
arc1.makegarch garch1
Cross-references
Test for autoregressive conditional heteroskedasticity (ARCH).
Carries out Lagrange Multiplier (LM) tests for ARCH in the residuals of a single least squares equation.
Syntax
eq_name.archtest(options)
Options
You must specify the order of ARCH for which you wish to test. The number of lags to be included in the test equation should be provided in parentheses after the arch keyword.
Other Options:
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
equation eq1.ls output c labor capital
eq1.archtest(4)
Regresses OUTPUT on a constant, LABOR, and CAPITAL, and tests for ARCH up to order 4.
equation eq1.arch sp500 c
eq1.archtest(4)
Estimates a GARCH(1,1) model with mean equation of SP500 on a constant and tests for additional ARCH up to order 4. Note that when performing an archtest as a view off of an estimated arch equation, EViews will use the standardized residuals (the residual of the mean equation divided by the estimated conditional standard deviation) to form the test.
Cross-references
See
“ARCH LM Test” for further discussion of testing ARCH and
“ARCH and GARCH Estimation” for a general discussion of working with ARCH models in EViews.
See also
Equation::hettest for a more full-featured version of this test.
Estimate an equation with autoregressive distributed lags using linear and nonlinear least squares or quantile regression.
Syntax
equation.ardl(options) linear_regs [@ static_regs] [@asy dual_asymmetric_regs] [@asylr long_run_asymmetric_regs] [@asysr short_run_asymmetric_regs]
The linear_regs specification is required:
• The linear_regs list should be the dependent variable followed by a list of linear distributed-lag regressors.
The remaining specifications are optional
• static_regs should be a list of static regressors, not including a constant or trend term.
• dual_asymmetric_regs are distributed-lag regressors which are asymmetric both in the short-run and long-run.
• long_run_asymmetric_regs regressors are distributed lag-regressors which are asymmetric in the long-run but symmetric in the short-run.
• short_run_asymmetric_regs are asymmetric regressors which are distributed lag-regressors which are asymmetric in the short-run but symmetric in the long-run.
You may specify the lag for an individual distributed-lag variable using the “@fl(variable, lag)” syntax. For instance, if the variable X should use 3 lags, irrespective of the fixed or automatic lag settings, you may specify this by entering “@fl(x, 3)” in the regressor list.
Options
Least Squares ARDL Options
method=arg (default = “ls”) | Set the method of estimation: "ls" (least-squares regression, default) or "qreg" (quantile regression). |
determ=arg (default = “rconst”) | Johansen deterministic trend type: “none” (no deterministics), “rconst” (restricted constant and no trend), “uconst” (unrestricted constant and no trend), “rtrend” (unrestricted constant and restricted trend, “utrend” (unrestricted constant and unrestricted trend). |
trend=arg (deprecated) | Johansen deterministic trend type: “none” (no deterministics), “const” (restricted constant and no trend, default), “uconst” (unrestricted constant and no trend), “linear” (unrestricted constant and restricted trend, “ulinear” (unrestricted constant and unrestricted trend). Note: this is a deprecated s option which handles a subset of cases covered by the “determ=” option |
fixed | Do not use automatic selection for lag lengths. This option must be used with the “deplags=” and “reglags=” options. |
deplags=int (default = 4) | Set the number of lags for the dependent variable to int. If automatic selection is used, this sets the maximum number of possible lags. If fixed lags are used (the fixed option is set), this fixes the number of lags. |
reglags=int (default = 4) | Set the number of lags for the explanatory variables (dynamic regressors) to int. If automatic selection is used, this sets the maximum number of possible lags. If fixed lags are used (the fixed option is set), this fixes the number of lags for each regressor. |
ic=key (default =“aic”) | Set the method of automatic model selection. key may take values of “aic” (Akaike information criterion, default), “bic” (Schwarz criterion), “hq” (Hannan-Quinn criterion) or “rbar2” (Adjusted R-squared, not applicable in panel workfiles). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “hac” (Newey-West HAC, available for nonlinear least squares or ARMA estimated by CLS).. |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 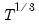 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Quantile ARDL Options
quant=number (default = 0.5) | Quantile to be fit (where number is a value between 0 and 1). |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=arg (default=“sandwich”) | Method for computing coefficient covariance matrix: “iid” (ordinary estimates), “sandwich” (Huber sandwich estimates), “boot” (bootstrap estimates). When “cov=iid” or “cov=sandwich”, EViews will use the sparsity nuisance parameter calculation specified in “spmethod=” when estimating the coefficient covariance matrix. |
bwmethod=arg (default = “hs”) | Method for automatically selecting bandwidth value for use in estimation of sparsity and coefficient covariance matrix: “hs” (Hall-Sheather), “bf” (Bofinger), “c” (Chamberlain). |
bw =number | Use user-specified bandwidth value in place of automatic method specified in “bwmethod=”. |
bwsize=number (default = 0.05) | Size parameter for use in computation of bandwidth (used when “bw=hs” and “bw=bf”). |
spmethod=arg (default=“kernel”) | Sparsity estimation method: “resid” (Siddiqui using residuals), “fitted” (Siddiqui using fitted quantiles at mean values of regressors), “kernel” (Kernel density using residuals) Note: “spmethod=resid” is not available when “cov=sandwich”. |
btmethod=arg (default= “pair”) | Bootstrap method: “resid” (residual bootstrap), “pair” (xy-pair bootstrap), “mcmb” (MCMB bootstrap), “mcmba” (MCMB-A bootstrap). |
btreps=integer (default=100) | Number of bootstrap repetitions |
btseed=positive integer | Seed the bootstrap random number generator. If not specified, EViews will seed the bootstrap random number generator with a single integer draw from the default global random number generator. |
btrnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator for the bootstrap: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
btobs=integer | Number of observations for bootstrap subsampling (when “bsmethod=pair”). Should be significantly greater than the number of regressors and less than or equal to the number of observations used in estimation. EViews will automatically restrict values to the range from the number of regressors and the number of estimation observations. If omitted, the bootstrap will use the number of observations used in estimation. |
btout=name | (optional) Matrix to hold results of bootstrap simulations. |
k=arg (default=“e”) | Kernel function for sparsity and coefficient covariance matrix estimation (when “spmethod=kernel”): “e” (Epanechnikov), “r” (Triangular), “u” (Uniform), “n” (Normal–Gaussian), “b” (Biweight–Quartic), “t” (Triweight), “c” (Cosinus). |
m=integer | Maximum number of iterations. |
s | Use the current coefficient values in estimator coefficient vector as starting values (see also
param). |
s=number (default =0) | Determine starting values for equations. Specify a number between 0 and 1 representing the fraction of preliminary least squares coefficient estimates. Note that out of range values are set to the default. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
opens example data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000.
The following command
equation eq01.ardl(deplags=8, reglags=8) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable, and the log of real GDP as a dynamic regressor. Quarterly dummy variables are included as static regressors. Automatic model selection is used to determine the number of lags of LOG(REALCONS) and LOG(REALGDP).
The command
equation eq02.ardl(deplags=3, reglags=3, fixed) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
estimates a second model, replicating Example 20.4 from Greene, with a fixed three lags of the dependent variable and three lags of the regressor.
equation eq03.ardl(deplags=1, reglags=1, fixed) log(realcons) log(realgdp) @asy log(realgovt)
The line above estimates an ARDL(1,1,1) model with the log of real consumption as the dependent variable, the log of real GDP as a linear regressor, and log of real government expenditures as a dual asymmetric regressor.
equation eq04.ardl(deplags=1, reglags=1, fixed) log(realcons) log(realgdp) @asy log(realgovt) @asysr log(realinvs)
extends the previous model and estimates an ARDL(1,1,1,1) model by including the log of real investments as a long-run asymmetric regressor.
equation eq05.ardl(deplags=1, reglags=1, fixed) log(realcons) log(realgdp) @asy log(realgovt) @asysr log(realinvs) @asylr log(tbilrate)
The line above extends the previous model and estimates an ARDL(1,1,1,1,1) model by including the log of treasury bill rates as a short-run asymmetric regressor.
wfopen oecd.wf1
equation eq06.ardl(fixed, deplags=1, reglags=1) log(cons) log(inf) log(inc)
This example estimates a panel ARDL model using the workfile “OECD.wf1”. This model replicates that given in the original Pesaran, Shin and Smith 1999 paper. Model selection is not used to choose the optimal lag lengths, rather a fixed single lag of both the dependent variable and the regressor is employed.
equation eq07.ardl(method=qreg, ls=fixed, deplags=1, reglags=1, quant=0.4) log(realcons) log(realgdp)
This command estimates a QARDL(1,1) model where lag selection is fixed for both the dependent and independent regressors, and the quantile value is 0.4.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
Examine ARMA structure of estimated equation.
Provides diagnostic graphical and tabular views that aid you in assessing the structure of the ARMA component of an estimated equation. The view is currently available only for equations specified by list and estimated by least squares that include at least one AR or MA term. There are four views types available: roots, correlogram, impulse response, and frequency spectrum.
Syntax
eq_name.arma(type=arg [,options])
where eq_name is the name of an equation object specified by list, estimated by least squares, and contains at least one ARMA term.
Options
type=arg | Required “type=” option selects the type of ARMA structure output: “root” displays the inverse roots of the AR/MA characteristic polynomials, “acf” displays the second moments (autocorrelation and partial autocorrelation) for the data in the estimation sample and for the estimated model, “imp” displays the impulse responses., “freq” displays the frequency spectrum. |
t | Displays the table view of the results for the view specified by the “type=” option. By default, EViews will display a graphical view of the ARMA results. |
hrz=arg | Specifies the maximum lag length for “type=acf”, and the maximum horizon (periods) for “type=imp”. |
imp=arg | Specifies the size of the impulse for the impulse response (“type=imp”) view. By default, EViews will use the regression estimated standard error. |
save=arg | Stores the results as a matrix object with the specified name. The matrix holds the results roughly as displayed in the table view of the corresponding type. For “type=root”, roots for the AR and MA polynomials will be stored in separate matrices as NAME_AR and NAME_MA, where “NAME” is the name given by the “save=” option. |
prompt | Force the dialog to appear from within a program. |
p | Print the table or graph output. |
Examples
eq1.arma(type=root, save=root)
displays and saves the ARMA roots from the estimated equation EQ1. The roots will be placed in the matrix object ROOT.
eq1.arma(type=acf, hrz=25, save=acf)
computes the second moments (autocorrelation and partial autocorrelations) for the observations in the sample and the estimated model. The results are computed for a 25 period horizon. We save the results in the matrix object ACF.
eq1.arma(type=imp, hrz=25, save=imp)
computes the 25 period impulse-response function implied by the estimated ARMA coefficients. EViews will use the default 1 standard error of the estimated equation as the shock, and will save the results in the matrix object IMP.
eq1.arma(type=freq)
displays the frequency spectrum in graph form.
Cross-references
Compute serial correlation LM (Lagrange multiplier) test.
Carries out Breusch-Godfrey Lagrange Multiplier (LM) tests for serial correlation in the estimation residuals.
Syntax
eq_name.auto(order, options)
You must specify the order of serial correlation for which you wish to test. You should specify the number of lags in parentheses after the auto keyword, followed by any additional options.
Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
To regress OUTPUT on a constant, LABOR, and CAPITAL, and test for serial correlation of up to order four you may use the commands:
equation eq1.ls output c labor capital
eq1.auto(4)
The commands:
output(t) c:\result\artest.txt
equation eq1.ls cons c y y(-1)
eq1.auto(12, p)
perform a regression of CONS on a constant, Y and lagged Y, and test for serial correlation of up to order twelve. The first line redirects printed tables/text to the ARTEST.TXT file.
Cross-references
See
“Serial Correlation LM Test” for further discussion of the Breusch-Godfrey test.
Estimate binary dependent variable models.
Estimates models where the binary dependent variable Y is either zero or one (probit, logit, gompit).
Syntax
eq_name.binary(options) y x1 [x2 x3 ...]
eq_name.binary(options) specification
Options
d=arg (default=“n”) | Specify likelihood: normal likelihood function, probit (“n”), logistic likelihood function, logit (“l”), Type I extreme value likelihood function, Gompit (“x”). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “glm” (GLM method), “cr” (cluster robust). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian - default). (Applicable when non-legacy “optmethod=”.) |
df | Degree-of-freedom correct the coefficient covariance estimate.(For non-cluster robust methods estimated using non-legacy estimation). |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option applicable when “optmethod=legacy”). |
g | GLM standard errors and covariances. (Legacy option applicable when “optmethod=legacy”). |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
To estimate a logit model of Y using a constant, WAGE, EDU, and KIDS, and computing Huber-White standard errors, you may use the command:
equation eq1.binary(d=l,cov=huber) y c wage edu kids
Note that this estimation uses the default global optimization options. The commands:
param c(1) .1 c(2) .1 c(3) .1
equation probit1.binary(s) y c x2 x3
estimate a probit model of Y on a constant, X2, and X3, using the specified starting values. The commands:
coef beta_probit = probit1.@coefs
matrix cov_probit = probit1.@coefcov
store the estimated coefficients and coefficient covariances in the coefficient vector BETA_PROBIT and matrix COV_PROBIT.
Cross-references
Perform the Pesaran, Shin and Smith (2001) bounds test of long-run relationships from an ARDL estimated equation.
This view displays a spool object with the ARDL bounds test diagnostics. The first table is a summary of the test along with statistic values. The second table summarizes the bound test critical values associated with the F-statistic. When appropriate (the deterministic case does not include a restricted constant (cases 3 and 5), a third table summarizes the bound test critical values associated with the t-statistic.
Syntax
eq_name.boundstest(options)
Options
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
equation eq02.ardl(deplags=3, reglags=3, fixed) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
show eq02.boundstest
This example uses data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000. The first line of this example downloads the data set, the second line creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable. Three lags of the dependent variable, and three lags of the log of real GDP are used as dynamic regressors. Quarterly dummy variables are included as static regressors.
The final line performs the Pesaran, Shin and Smith (2001) bounds test to test for a long-run relationship between the log of real consumption and the log of real GDP.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
Estimation by linear least squares regression with breakpoints.
Syntax
eq_name.breakls(options) y z1 [z2 z3 ...] [@nv x1 x2 x3 ...]
List the dependent variable first, followed by a list of the independent variables that have coefficients which are allowed to vary across breaks, followed optionally by the keyword @nv and a list of non-varying coefficient variables.
Options
Breakpoint Options
method=arg (default=“seqplus1”) | Breakpoint selection method: “seqplus1” (sequential tests of single 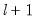 versus 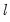 breaks), “seqall” (sequential test of all possible 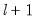 versus 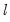 breaks), “glob” (tests of global 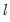 vs. no breaks), “globplus1” (tests of 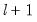 versus 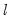 globally determined breaks), “globinfo” (information criteria evaluation),“user” (user-specified break dates). |
select=arg | Sub-method setting (options depend on “method=”). (1) if “method=glob”: Sequential (“seq”) (default), Highest significant (“high”), 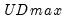 (“udmax”), 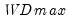 (“wdmax”). (2) if “method=globinfo”: Schwarz criterion (“bic” or “sic”) (default), Liu-Wu-Zidek criterion (“lwz”). |
trim=arg (default=5) | Trimming percentage for determining minimum segment size (5, 10, 15, 20, 25). |
maxbreaks=integer (default=5) | Maximum number of breakpoints to allow (not applicable if “method=seqall”). |
maxlevels=integer (default=5) | Maximum number of break levels to consider in sequential testing (applicable when “method=sequall”). |
breaks="arg" | User-specified break dates entered in double quotes. For use when “method=user”. |
size=arg (default=5) | Test sizes for use in sequential determination and final test evaluation (10, 5, 2.5, 1) corresponding to 0.10, 0.05, 0.025, 0.01, respectively |
heterr | Assume regimes specific error distributions in variance computation. |
commondata | Assume a common distribution for the data across segments (only applicable if original equation is estimated with a robust covariance method, “heterr” is not specified). |
General Options
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=keyword | Covariance type (optional): “white” (White diagonal matrix), “hac” (Newey-West HAC). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 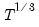 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
covbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
coef=arg | Specify the name of the coefficient vector; the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
equation eq1.breakls m1 c unemp
uses the Bai-Perron sequential
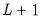
versus
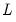
tests to determine the optimal breaks in a model regressing M1 on the breaking variables C and UNEMP.
equation eq2.breakls(method=glob, select=high) m1 c unemp
uses the global Bai-Perron
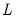
versus none tests to determine the breaks. The selected break will be the highest significant number of breaks.
equation eq3.breakls(size=5, trim=10) m1 c unemp
lowers the sequential test size from 0.10 to 0.05, and raises the trimming to 10 percent.
equation eq4.breakls(method=user, breaks="1990q1 2010q4") m1 c @nv unemp
estimates the model with two user-specified break dates. In addition, the variable UNEMP is restricted to have common coefficients across the regimes.
Cross-references
See
Equation::multibreak for multiple breakpoint testing.
Display the breakpoint specification results for an equation estimated using breakls.
Syntax
eq_name.breakspec
Options
p | Print basic estimation results. |
Examples
equation eq1.breakls m1 c unemp
eq1.breakspec(p)
displays and prints the breakpoint determination results for the equation EQ1 estimated using Bai-Perron sequential
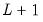
versus
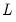
tests to determine the optimal breaks.
Cross-references
Breakpoint test.
Carries out a breakpoint test for parameter stability in equations estimated using TSLS and GMM.
See chow for related tests in equations estimated using least squares.
Syntax
eq_name.breaktest obs1 [obs2 obs3....]
You must provide the breakpoint observations (using dates or observation numbers) to be tested. To specify more than one breakpoint, separate the breakpoints by a space.
Examples
The commands
equation eq1.gmm m1 c gdp cpi @ gdp(-1) cpi(-1)
eq1.breaktest 1960 1970
perform a GMM estimation of M1 on a constant, GDP and CPI, with lagged values of GDP and CPI used as instruments, and then perform a breakpoint test to test whether the parameter estimates for the periods prior to 1960, during the 1960s, and then after 1970 are stable.
Cross-references
Test for the presence of cross-sectional dependence in the residuals of panels equations.
Computes the Breusch-Pagan (1980) LM, Pesaran (2004) scaled LM, Pesaran (2004) CD, and Baltagi, and Feng and Kao (2012) bias-corrected scaled LM test for the residuals of a panel or pool equation, or panel series.
Syntax
eq_name.cdtest
Options
Examples
equation eq1.ls(cx=f) @log(gsp) c @log(p_cap) @log(pc) @log(emp) unemp
eq1.cdtest
will estimate a panel model using the fixed effect estimator (EQ1) and then will compute and display the panel residual dependence test results.
Cross-references
Confidence ellipses for coefficient restrictions.
The cellipse view displays confidence ellipses for pairs of coefficient restrictions for an equation object.
Syntax
eq_name.cellipse(options) restrictions
Enter the equation name, followed by a period, and the keyword cellipse. This should be followed by a list of the coefficient restrictions. Joint (multiple) coefficient restrictions should be separated by commas.
Options
ind=arg | Specifies whether and how to draw the individual coefficient intervals. The default is “ind=line” which plots the individual coefficient intervals as dashed lines. “ind=none” does not plot the individual intervals, while “ind=shade” plots the individual intervals as a shaded rectangle. |
size= number (default=0.95) | Set the size (level) of the confidence ellipse. You may specify more than one size by specifying a space separated list enclosed in double quotes. |
dist= arg | Select the distribution to use for the critical value associated with the ellipse size. The default depends on estimation object and method. If the parameter estimates are least-squares based, the 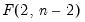 distribution is used; if the parameter estimates are likelihood based, the 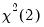 distribution will be employed. “dist=f” forces use of the F-distribution, while “dist=c” uses the 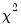 distribution. |
prompt | Force the dialog to appear from within a program. |
p | Print the graph. |
Examples
The two commands:
eq1.cellipse c(1), c(2), c(3)
eq1.cellipse c(1)=0, c(2)=0, c(3)=0
both display a graph showing the 0.95-confidence ellipse for C(1) and C(2), C(1) and C(3), and C(2) and C(3).
eq1.cellipse(dist=c,size="0.9 0.7 0.5") c(1), c(2)
displays multiple confidence ellipses (contours) for C(1) and C(2).
Cross-references
Estimation of censored and truncated models.
Estimates models where the dependent variable is either censored or truncated. The allowable specifications include the standard Tobit model.
Syntax
eq_name.censored(options) y x1 [x2 x3]
eq_name.censored(options) specification
Options
l=number (default=0) | Set value for the left censoring limit. |
r=number (default=none) | Set value for the right censoring limit. |
l=series_name, i | Set series name of the indicator variable for the left censoring limit. |
r=series_name, i | Set series name of the indicator variable for the right censoring limit. |
t | Estimate truncated model. |
d=arg (default=“n”) | Specify error distribution: normal (“n”), logistic (“l”), Type I extreme value (“x”). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich methods)., “cr” (cluster robust). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian - default). (Applicable when non-legacy “optmethod=”). |
df | Degree-of-freedom correct the coefficient covariance estimate.(For non-cluster robust methods estimated using non-legacy estimation). |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option applicable when “optmethod=legacy”). |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
The command:
eq1.censored(cov=huber) hours c wage edu kids
estimates a censored regression model of HOURS on a constant, WAGE, EDU, and KIDS with QML standard errors. This command uses the default normal likelihood, with left-censoring at HOURS=0, no right censoring, and the quadratic hill climbing algorithm.
Cross-references
See
“Discrete and Limited Dependent Variable Models” for discussion of censored and truncated regression models.
Chow test for stability.
Carries out Chow breakpoint or Chow forecast tests for parameter constancy.
Syntax
eq_name.chow(options) obs1 [obs2 obs3 ...] @ x1 x2 x3
You must provide the breakpoint observations (using dates or observation numbers) to be tested. To specify more than one breakpoint, separate the breakpoints by a space. For the Chow breakpoint test, if the equation is specified by list and contains no nonlinear terms, you may specify a subset of the regressors to be tested for a breakpoint after an “@” sign.
Options
f | Chow forecast test. For this option, you must specify a single breakpoint to test (default performs breakpoint test). |
p | Print the result of test. |
Examples
The commands:
equation eq1.ls m1 c gdp cpi ar(1)
eq1.chow 1970Q1 1980Q1
perform a regression of M1 on a constant, GDP, and CPI with first order autoregressive errors, and employ a Chow breakpoint test to determine whether the parameters before the 1970’s, during the 1970’s, and after the 1970’s are “stable”.
To regress the log of SPOT on a constant, the log of P_US, and the log of P_UK, and to carry out the Chow forecast test starting from 1973, enter the commands:
equation ppp.ls log(spot) c log(p_us) log(p_uk)
ppp.chow(f) 1973
To test whether only the constant term and the coefficient on the log of P_US prior to and after 1970 are “stable” enter the commands:
ppp.chow 1970 @ c log(p_us)
Cross-references
See
“Chow's Breakpoint Test” for further discussion.
Confidence interval.
The confidence interval view displays a table of confidence intervals for each of the coefficients in the equation.
Syntax
eq_name.cinterval(options) arg
where arg is a list of confidence levels, or the name of a scalar or vector in the workfile containing confidence levels.
Options
prompt | Force the dialog to appear from within a program. |
nopair | Display the intervals concentrically. The default is to display them in pairs for each probability value |
Examples
The set of commands:
equation eq1.ls lwage c edu edu^2 union
eq1.cinterval .95 .9 .75
displays the 95% confidence intervals followed by the 90% confidence levels, followed by the 75% confidence levels.
eq1.cinterval(nopair) .95 .9 .75
displays the 75% confidence interals nested inside the 90% intervals which in turn are nested inside the 95% intervals.
Cross-references
Clear the contents of the history attribute for equation objects.
Removes the equation’s history attribute, as shown in the label view of the equation.
Syntax
equation_name.clearhist
Examples
eq1.clearhist
eq1.label
The first line removes the history from the equation EQ1, and the second line displays the label view of EQ1, including the now blank history field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Clear the contents of the remarks attribute.
Removes the equations’s remarks attribute, as shown in the label view of the equation.
Syntax
equation_name.clearremarks
Examples
e1.clearremarks
e1.label
The first line removes the remarks from the equation E1, and the second line displays the label view of E1, including the now blank remarks field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Coefficient covariance matrix.
Displays the covariances of the coefficient estimates for an estimated equation.
Syntax
eq_name.coefcov(options)
Options
p | Print the coefficient covariance matrix. |
Examples
The set of commands:
equation eq1.ls lwage c edu edu^2 union
eq1.coefcov
declares and estimates equation EQ1 and displays the coefficient covariance matrix in a window. To store the coefficient covariance matrix as a sym object, use “@coefcov”:
sym eqcov = eq1.@coefcov
Cross-references
Displays the coefficients associated with variables in the equation specification.
Syntax
equation_name.coeflabel(options)
Options
Examples
equation eq1.ls m1 c inc tb3 ar(1)
eq1.coeflabel(p)
displays and prints a view showing the coefficients associated with each variable in the equation specification.
Cross-references
Display the matrix of lambda and coefficients for elastic net, ridge, and Lasso models.
Syntax
eq_name.coefmatrix(options)
Options
Cross-references
For further discussion, see
“Elastic Net and Lasso”.
Display graphs of the paths of the coefficients plotted against lambda, fit measures, and estimation values.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
You may plot the coefficient values against the paths of: the penalty parameter lambda, R-squared, adjusted R-square, standard error of the regression, sum-of-squared residuals, L1-norm coefficient penalty, L2-norm squared coefficient penalty, and the estimation objective.
A vertical line will be included to identify the selected optimal lambda.
Only coefficients that have non-zero values for at least one lambda in the path will be displayed.
By default, EViews will display a spool object containing all of the plots. You may use the “type=” option to produce a specific graph.
Syntax
eq_name.coefpaths(options)
Options
type=arg | Graph of coefficient against the path of: “lambda” (log lambda), “r2” (R-squared), “rbar2” (Adjusted R-squared), “se” (standard error of regression), “ssr” (sum-of-squared residuals), “l1” (L1 coefficient penalty, if applicable), “l2” (L2-squared coefficient penalty, if applicable), “estobj” (estimation objective). If “type=” is not provided, EViews will display the spool object all of the graphs. |
p | Print output. |
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.coefpath
displays estimates a spool containing graphs of all of the coefficients plotted against the paths of lambda, fit measures, and estimation measures.
my_eq.coefpath(type=lambda, p)
displays and prints a single graph of the coefficient lambda path, while
my_eq_coefpath(type=r2)
plots the coefficients against the path of the R-squared.
Cross-references
For further discussion, see
“Elastic Net and Lasso”The data underlying these graphs are available via the data members @lambdacoefs, @lambdaest and @lambdafit.
Scaled coefficients.
Displays the coefficient estimates, the standardized coefficient estimates and the elasticity at means.
Syntax
eq_name.coefscale
Examples
The set of commands:
equation eq1.ls lwage c edu edu^2 union
eq1.coefscale
produces the coefficient scale table view of EQ1.
Cross-references
Test for cointegration between series in an equation.
Test for cointegration between series in an equation estimated by
Equation::cointreg. You may perform a Hansen Instability Test, Park Added Variable (Spurious Trends) Test, or between a residual-based Engle-Granger or Phillips-Ouliaris test.
Johansen tests for cointegration may be performed from a group or a VAR object (see
Group::coint and
Var::coint).
The cointegrating equation specification is taken from the equation. Additional test specification components are specified as options and arguments.
Syntax
Equation View: eq_name.coint(options) [arg]
where
method=arg (default=“hansen”) | Test method: Hansen’s Instability test (“hansen”), Park’s Added Variable (“park”), Engle-Granger residual test (“eg”), Phillips-Ouliaris residual test (“po”). |
and arg is an optional list describing additional regressors to include in the Park Added Regressors test (when “method=park” is specified).
The Park, Engle-Granger, and Phillips-Ouliaris tests all have options which control various aspects of the test.
Options
Options for the Park Test
The following option, along with the optional argument described above, determines the additional regressors to include in the test equation.
trend=arg (default=two orders higher than trend in estimated equation) | Specification for the powers of trend to include in the test equation: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”), Cubic trend (“cubic”), Quartic trend (“quartic”), integer (user-specified power). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. Only trend orders higher than those specified in the original equation will be considered. |
p | Print results. |
Options for the Engle-Granger Test
The following options determine the specification of the Engle-Granger test (Augmented Dickey-Fuller) equation and the calculation of the variances used in the test statistic.
lag=arg (default=“a”) | Method of selecting the lag length (number of first difference terms) to be included in the regression: “a” (automatic information criterion based selection), or integer (user-specified lag length). |
lagtype=arg (default=“sic”) | Information criterion or method to use when computing automatic lag length selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “msaic” (Modified Akaike), “msic” (Modified Schwarz), “mhqc” (Modified Hannan-Quinn), “tstat” (t-statistic). |
maxlag=integer | Maximum lag length to consider when performing automatic lag-length selection default=  where  is the number of coefficients in the cointegrating equation. Applicable when “lag=a”. |
lagpval=number (default=0.10) | Probability threshold to use when performing automatic lag-length selection using a t-test criterion. Applicable when both “lag=a” and “lagtype=tstat”. |
nodf | Do not degree-of-freedom correct estimates of the variances. |
p | Print results. |
Options for the Phillips-Ouliaris Test
The following options control the computation of the symmetric and one-sided long-run variances in the Phillips-Ouliaris test.
Basic Options
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
p | Print results. |
HAC Whitening Options
lag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Kernel Options
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
bw=arg (default=“nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
bwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Examples
Hansen
equation base_eq.cointreg(trend=linear, bw=andrews, kern=quadspec)
base_eq.coint
estimates the cointegrating equation BASE_EQ using FMOLS and performs the Hansen cointegration test.
Park
base_eq.coint(method=park)
conducts the default Park test, which for BASE_EQ involves testing the significance of the quadratic and cubic trend coefficients.
base_eq.coint(method=park, trend=quartic) mytrend
performs a test which evaluates the significance of the quadratic, cubic, and quartic terms, and user trend variable MYTREND.
base_eq.coint(method=eg, trend=6)
estimates the test equation with trend powers up to 6.
Engle-Granger
base_eq.coint(method=eg)
performs the default Engle-Granger test using SIC and an observation-based maximum number of lags to determine the lags for an ADF equation.
base_eq.coint(method=eg, lag=a, lagtype=tstat, lagpval=.15, maxlag=10)
uses a sequential t-test starting at lag 10 with threshold probability 0.15 to determine the number of lags.
base_eq.coint(method=eg, lag=5)
conducts an Engle-Granger cointegration test with a fixed lag of 5.
Phillips-Ouliaris
base_eq.coint(method=po)
performs the default Phillips-Ouliaris test using a Bartlett kernel and Newey-West fixed bandwidth.
base_eq.coint(method=po, bw=andrews, kernel=quadspec, nodf)
estimates the long-run covariances using a Quadratic Spectral kernel, Andrews automatic bandwidth, and no degrees-of-freedom correction.
base_eq.coint(method=po, lag=1, bw=4)
constructs the long-run covariances using AR(1) prewhitening, a fixed bandwidth of 4, and the Bartlett kernel.
Cross-references
See
“Cointegration Testing”. See also
Group::coint for testing from a group object.
View a graph of the estimated cointegrating relation form of an ARDL estimated equation.
This view is only available for non-panel equations estimated using the ARDL method.
Syntax
equation_name.cointgraph
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
equation eq02.ardl(deplags=3, reglags=3, fixed) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
show eq02.cointgraph
This example uses data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000. The first line of this example downloads the data set, the second line creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable. Three lags of the dependent variable, and three lags of the log of real GDP are used as dynamic regressors. Quarterly dummy variables are included as static regressors.
The final line views a graph of the cointegration representation of the estimation.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
Estimate a cointegrating equation using Fully Modified OLS (FMOLS), Canonical Cointegrating Regression (CCR), or Dynamic OLS (DOLS) in single time series settings, and Panel FMOLS and DOLS in panel workfiles.
Syntax
eq_name.cointreg(options) y x1 [x2 x3 ...] [@determ determ_spec] [@regdeterm regdeterm_spec]
List the cointreg keyword, followed by the dependent variable and a list of the cointegrating variables.
Cointegrating equation specifications that include a constant, linear, or quadratic trends, should use the “trend=” option to specify those terms. If any of those terms are in the stochastic regressors equations but not in the cointegrating equation, they should be specified using the “regtrend=” option.
Deterministic trend regressors that are not covered by the list above may be specified using the keywords @determ and @regdeterm. To specify deterministic trend regressors that enter into the regressor and cointegrating equations, you should add the keyword @determ followed by the list of trend regressors. To specify deterministic trends that enter in the regressor equations but not the cointegrating equation, you should include the keyword @regdeterm followed by the list of trend regressors.
Basic Options
method=arg (default=“fmols”) | Estimation method: Fully Modified OLS (“fmols”), Canonical Cointegrating Regression (“ccr”), Dynamic OLS (“dols”) Note that CCR estimation is not available in panel settings. |
trend=arg (default=“const”) | Specification for the powers of trend to include in the cointegrating and regressor equations: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regtrend=arg (default=“none”) | Additional trends to include in the regressor equations (but not the cointegrating equation): None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Only trend orders higher than those specified by “trend=” will be considered. Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regdiff | Estimate the regressor equation innovations directly using the difference specifications. |
coef=arg | Specify the name of the coefficient vector; the default behavior is to use the “C” coefficient vector. |
btwcoefs=arg | Save the cross-section specific deterministic coefficient estimates in a matrix object (one row per cross-section). |
btwcovs=arg | Save the covariances of the cross-section specific deterministic coefficient estimates in a matrix object (one row per cross-section, with each row holding the vech of the covariance). |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
In addition to these options, there are specialized options for each estimation method.
Panel Options
panmethod=arg (default=“pooled”) | Panel estimation method: pooled (“pooled”), pooled weighted (“weighted”), grouped (“grouped”) |
pancov=sandwich | Estimate the coefficient covariance using a sandwich method that allows for cross-section heterogeneity. |
Options for FMOLS and CCR
To estimate FMOLS or CCR use the “method=fmols” or “method=ccr” options. The following options control the computation of the symmetric and one-sided long-run covariance matrices and the estimate of the coefficient covariance.
HAC Whitening Options
lag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Kernel Options
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
bw=arg (default=“nwfixed”) | Bandwidth:: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
bwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Coefficient Covariance
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
Panel Options
hetfirst | Estimate the first-stage regression assuming heterogeneous coefficients. For FMOLS specifications estimated using pooled or pooled weighted methods (“panmethod =pooled”, “panmethod=weighted”) |
Options for DOLS
To estimate using DOLS use the “method=dols” option. The following options control the specification of the lags and leads and the estimate of the coefficient covariance.
lltype=arg (default=“fixed”) | Lag-lead method: fixed values (“fixed”), automatic selection - Akaike (“aic”), automatic - Schwarz (“sic”), automatic - Hannan-Quinn (“hqc”), None (“none”). |
lag=arg | Lag specification: integer (required user-specified number of lags if “lltype=fixed”). |
lead=arg | Lead specification: integer (required user-specified number of lags if “lltype=fixed”). |
maxll=integer | Maximum lag and lead-length for automatic selection (optional user-specified integer if “lltype=” is used to specify automatic selection). The default is an observation-based maximum. |
cov=arg | Coefficient covariance method: (default) long-run variance scaled OLS, unscaled OLS (“ols”), White (“white”), Newey-West (“hac”). |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
For the default covariance calculation or “cov=hac”, the following options control the computation of the long-run variance or robust covariance:
HAC Covariance Whitening Options (if default covariance or “cov=hac”)
covlag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Covariance Kernel Options (if default covariance or “cov=hac”)
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “covbw=neweywest”). |
covbwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
covbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Panel Options
Weighted coefficient or coefficient covariance estimation in panel DOLS requires individual estimates of the long-run variances of the residuals. You may compute these estimates using the standard default long-run variance options, or you may choose to estimate it using the ordinary variance.
For weighted estimation we have:
panwgtlag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
panwgtinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lrvarlag=a”). |
panwgtmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lrvarlag=a”). The default is an observation-based maximum. |
panwgtkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
panwgtbw=arg (default=“nwfixed”) | Bandwidth:: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
panwgtnwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
panwgtbwoffset=integer (default=0) | Apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
panwgtbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
For the coefficient covariance estimation we have:
lrvar=ordinary | Compute DOLS estimates of the long-run residual variance used in covariance calculation using the ordinary variance. |
lrvarlag=arg (default=0) | For DOLS estimates of the long-run residual variance used in covariance calculation, lag specification: integer (user-specified lag value), “a” (automatic selection). |
lrvarinfosel=arg (default=“aic”) | For DOLS estimates of the long-run residual variance used in covariance calculation, information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lrvarlag=a”). |
lrvarmaxlag=integer | For DOLS estimates of the long-run residual variance used in covariance calculation, maximum lag-length for automatic selection (optional) (if “lrvarlag=a”). The default is an observation-based maximum. |
lrvarkern=arg (default=“bart”) | For DOLS estimates of the long-run residual variance used in covariance calculation, Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
lrvarbw=arg (default=“nwfixed”) | For DOLS estimates of the long-run residual variance used in covariance calculation, bandwidth:: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
lrvarnwlag=integer | For DOLS estimates of the long-run residual variance used in covariance calculation, Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
lrvarbwoffset=integer (default=0) | For DOLS estimates of the long-run residual variance used in covariance calculation, apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
lrvarbwint | For DOLS estimates of the long-run residual variance used in covariance calculation, use integer portion of bandwidth. |
Examples
FMOLS and CCR
To estimate, by FMOLS, the cointegrating equation for LC and LY including a constant, you may use:
equation fmols.cointreg(nodf, bw=andrews) lc ly
The long-run covariances are estimated nonparametrically using a Bartlett kernel and a bandwidth determined by the Andrews automatic selection method. The coefficient covariances are estimated with no degree-of-freedom correction.
To include a linear trend term in a model where the long-run covariances computed using the Quadratic Spectral kernel and a fixed bandwidth of 10, enter:
equation fmols.cointreg(trend=linear, nodf, bw=10, kern=quadspec) lc ly
A model with a cubic trend may be estimated using:
equation fmols.cointreg(trend=linear, lags=2, bw=neweywest, nwlag=10, kernel=parzen) lc ly @determ @trend^3
Here, the long-run covariances are estimated using a VAR(2) prewhitened Parzen kernel with Newey-West nonparametric bandwidth determined using 10 lags in the autocovariance calculations.
equation fmols.cointreg(trend=quadratic, bw=andrews, lags=a, infosel=aic, kernel=none, regdiff) lc ly @regdeterm @trend^3
estimates a restricted model with a cubic trend term that does not appear in the cointegrating equation using a parametric VARHAC with automatic lag length selection based on the AIC. The residuals for the regressors equations are obtained by estimating the difference specification.
To estimate by CCR, we provide the “method=ccr” option. The command
equation ccr.cointreg(method=ccr, lag=2, bw=andrews, kern=quadspec) lc ly
estimates, by CCR, the constant only model using a VAR(2) prewhitened Quadratic Spectral and Andrews automatic bandwidth selection.
equation ccr.cointreg(method=ccr, trend=linear, lag=a, maxlag=5, bw=andrews, kern=quadspec) lc ly
modifies the previous estimates by adding a linear trend term to the cointegrating and regressors equations, and changing the VAR prewhitening to automatic selection using the default SIC with a maximum lag length of 5.
equation ccr.cointreg(method=ccr, trend=linear, regtrend=quadratic, lag=a, maxlag=5, bw=andrews) lc ly
adds a quadratic trend term to the regressors equations only, and changes the kernel to the default Bartlett.
DOLS
equation dols.cointreg(method=dols, trend=linear, nodf, lag=4, lead=4) lc ly
estimates the linear specification using DOLS with four lags and leads. The coefficient covariance is obtained by rescaling the no d.f.-correction OLS covariance using the long-run variance of the residuals computed using the default Bartlett kernel and default fixed Newey-West bandwidth.
equation dols.cointreg(method=dols, trend=quadratic, nodf, lag=4, lead=2, covkern=bohman, covbw=10) lc ly @determ @trend^3
estimates a cubic specification using 4 lags and 2 leads, where the coefficient covariance uses a Bohman kernel and fixed bandwidth of 10.
equation dols.cointreg(method=dols, trend=quadratic, nodf, lag=4, lead=2, cov=hac, covkern=bohman, covbw=10) lc ly @determ @trend^3
estimates the same specification using a HAC covariance in place of the scaled OLS covariance.
equation sols.cointreg(method=dols, trend=quadratic, lltype=none, cov=ols) lc ly @determ @trend^3
computes the static OLS estimates with the usual OLS d.f. corrected coefficient covariance.
Cross-references
See
“Cointegrating Regression” for a discussion of single equation cointegrating regression. See
“Panel Cointegration Estimation” for discussion of estimation in panel settings.
See
“Technical Discussion” for a discussion of VEC estimation.
Display information about the cointegrating relation specification and the coefficients in an ARDL estimated equation.
Syntax
eq_name.cointrel(options)
Options
Example
ardl_eq.cointrel
displays a spool object with the table and graph showing the cointegrating relation.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
View the estimated cointegration form and the long-run coefficients table of an ARDL estimated equation.
This view is only available for non-panel equations estimated using the ARDL method.
Syntax
equation_name.cointrep
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
equation eq02.ardl(deplags=3, reglags=3, fixed) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
show eq02.cointrep
This example uses data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000. The first line of this example downloads the data set, the second line creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable. Three lags of the dependent variable, and three lags of the log of real GDP are used as dynamic regressors. Quarterly dummy variables are included as static regressors.
The final line views the cointegration representation of the estimation, as well as the long-run form of the coefficient estimates.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
Creates a copy of the equation.
Creates either a named or unnamed copy of the equation.
Syntax
equation_name.copy
equation_name.copy dest_name
Examples
eq1.copy
creates an unnamed copy of the equation EQ1.
eq1.copy eq2
creates EQ2, a copy of the equation EQ1.
Cross-references
Display autocorrelation and partial correlations.
Displays the correlogram and partial correlation functions of the residuals of the equation, together with the Q-statistics and p-values associated with each lag.
Syntax
eq_name.correl(n, options)
You must specify the largest lag n to use when computing the autocorrelations.
Options
Examples
eq1.correl(24)
Displays the correlograms of the residuals of EQ1 for up to 24 lags.
Cross-references
See
“Autocorrelations (AC)” and
“Partial Autocorrelations (PAC)” for a discussion of autocorrelation and partial correlation functions, respectively.
Correlogram of squared residuals.
Displays the autocorrelation and partial correlation functions of the squared residuals from an estimated equation, together with the Q-statistics and p-values associated with each lag.
Syntax
equation_name.correlsq(n, options)
You must specify the largest lag n to use when computing the autocorrelations.
Options
Examples
eq1.correlsq(24)
displays the correlograms of the squared residuals of EQ1 up to 24 lags.
Cross-references
See
“Autocorrelations (AC)” and
“Partial Autocorrelations (PAC)” for a discussion of autocorrelation and partial correlation functions, respectively.
Estimates models where the dependent variable is a nonnegative integer count.
Syntax
eq_name.count(options) y x1 [x2 x3...]
eq_name.count(options) specification
Follow the count keyword by the name of the dependent variable and a list of regressors or provide a linear specification.
Options
d=arg (default=“p”) | Likelihood specification: Poisson likelihood (“p”), normal quasi-likelihood (“n”), exponential likelihood (“e”), negative binomial likelihood or quasi-likelihood (“b”). |
v=positive_num (default=1) | Specify fixed value for QML parameter in normal and negative binomial quasi-likelihoods. |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich methods)., “glm” (GLM method),““cr” (cluster robust). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
df | Degree-of-freedom correct the coefficient covariance estimate.(For non-cluster robust methods estimated using non-legacy estimation). |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
g | GLM standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of the EViews default values (out of range values are set to “s=1”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
The command:
equation eq1.count(d=n,v=2,cov=glm) y c x1 x2
estimates a normal QML count model of Y on a constant, X1, and X2, with fixed variance parameter 2, and GLM standard errors.
equation eq1.count arrest c job police
eq1.makeresids(g) res_g
estimates a Poisson count model of ARREST on a constant, JOB, and POLICE, and stores the generalized residuals in the series RES_G.
equation eq1.count(d=p) y c x1
eq1.fit yhat
estimates a Poisson count model of Y on a constant and X1, and saves the fitted values (conditional mean) in the series YHAT.
equation eq1.count(d=p, h) y c x1
estimates the same model with QML standard errors and covariances.
Cross-references
See
“Count Models” for additional discussion.
Displays the coefficient covariance decomposition table.
Syntax
equation_name.cvardecomp
Examples
equation e1.ls y c x
eq1.cvardecomp
creates and estimates an equation named E1, and then displays the coefficient covariance decomposition table.
Cross-references
Display a graph of the cross-validation objective against the lambda path.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
Show a graph of the cross-validation objective for each value of lambda, along with bars representing +/– one standard deviation of the mean (for cross-validation methods that produce multiple training samples for each lambda).
A vertical line will be included to identify the selected optimal lambda.
Syntax
eq_name.cvgraph(options)
Options
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.cvgraph
displays a graph of the cross-validation measures of fit against the path of log lambda. The cross-validation selected optimal value of lambda is marked by a vertical line.
Cross-references
For further discussion, see
“Elastic Net and Lasso”The data underlying this graph are available via the data members @lambdafit, and @cvobjective.
Dependent variable frequency table.
Displays the frequency table for the dependent variable in binary, count, and ordered equations.
Syntax
equation_name.depfreq(options)
Options
p | Print the frequency table. |
Examples
eq1.depfreq(p)
displays and prints the dependent variable frequency.
Cross-references
Examine derivatives of the equation specification.
Display information about the derivatives of the equation specification in tabular, graphical, or summary form.
The (default) summary form shows information about how the derivative of the equation specification was computed, and will display the analytic expression for the derivative, or a note indicating that the derivative was computed numerically.
You may optionally choose a tabular or graphical display of the derivatives. The tabular form shows a spreadsheet view of the derivatives of the regression specification with respect to each coefficient (for each observation). The graphical form of the view shows this information in a multiple line graph.
Syntax
equation_name.derivs(options)
Options
t | Display spreadsheet view of the values of the derivatives with respect to the coefficients evaluated at each observation. |
g | Display multiple graphs showing the derivatives of the equation specification with respect to the coefficients, evaluated at each observation. |
p | Print results. |
Note that the “g” and “t” options may not be used at the same time.
Examples
To show a table view of the derivatives:
eq1.derivs(t)
To display and print the summary view:
eq1.derivs(p)
Cross-references
See
“Derivative Computation” for details on the computation of derivatives.
See also
Equation::makederivs for additional routines for examining derivatives, and
Equation::grads, and
Equation::makegrads for corresponding routines for gradients.
Estimate a equation in a panel structured workfile using the difference-in-difference estimator.
Syntax
equation.did(options) y [x1] [@ treatment]
List the dependent variable, followed by an optional list of exogenous regressors, followed by an “@” and then the binary treatment variable. You should not include a constant in the specification.
Options
coef=arg | Specify the name of the coefficient vector. The default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eq1.did asmrs @ post
estimates an equation by difference-in-difference with ASMRS as the outcome variable, and POST as the treatment variable.
equation eq2.did lemp lpop @ treated
estimates an equation by difference-in-difference with LEMP as the outcome variable, TREATED as the treatment variable, and LPOP as an exogenous regressor.
Cross-references
See
“Difference-in-Difference Estimation” for a discussion of difference-in-difference models.
Display the Callaway-Sant’Anna decomposition for difference-in-difference estimation.
For panel equations estimated using the difference-in-difference method.
Syntax
eq_name.didcs(options) [additional_regs]
You should follow the didcs keyword by an optional list of additional regressors added to the Callaway-Sant’Anna estimation.
Options
notyet | Use observations where an individual is not yet treated as the comparison group. Default is to only use individuals that are never treated as the comparison. |
both | Use both observations where an individual is never treated or has not yet been treated as the comparison group. Default is to only use individuals that are never treated as the comparison. |
p | Print output. |
Example
equation eq1.did lemp @ treated
eq1.didcs lpop
estimates an equation by difference-in-difference with LEMP as the outcome variable, and TREATED as the treatment variable, and then displays the Callaway-Sant’Anna decomposition, adding LPOP as an additional exogenous regressor.
Cross-references
See
“Difference-in-Difference Estimation” for a discussion of difference-in-difference models.
Display the Goodman-Bacon decomposition for difference-in-difference estimation.
For panel equations estimated using the difference-in-difference method.
Syntax
eq_name.didgbdecomp(options)
Options
Example
equation eq1.did asmrs @ post
eq1.didgbdecomp
estimates an equation by difference-in-difference with ASMRS as the outcome variable, and POST as the treatment variable, and then displays the Goodman-Bacon decomposition.
Cross-references
See
“Difference-in-Difference Estimation” for a discussion of difference-in-difference models.
Create an equation object with the underlying fixed-effects estimation of a difference-in-difference equation.
For panel equations estimated using the difference-in-difference method.
Syntax
eq_name.didmakeeq new_eqname
You should follow the didmakeeq keyword with the name of the new estimated equation with an equivalent specification to be created in the workfile.
Example
equation eq1.did asmrs @ post
eq1.didmakeeq eq_underlying
estimates an equation by difference-in-difference with ASMRS as the outcome variable, and POST as the treatment variable, and then creates the underlying fixed effects estimation in the equation object EQ_UNDERLYING
Cross-references
See
“Difference-in-Difference Estimation” for a discussion of difference-in-difference models.
Display difference-in-difference trends summary in graph or tabular form.
Syntax
eq_name.didtrends(options)
For panel equations estimated using the difference-in-difference method.
Options
t | Display results in a table. |
p | Print output. |
Example
equation eq1.did asmrs @ post
eq1.didtrends
eq1.didtrends(t)
estimates an equation by difference-in-difference with ASMRS as the outcome variable, and POST as the treatment variable, and then displays the trend summary graph, then as a table.
Cross-references
See
“Difference-in-Difference Estimation” for a discussion of difference-in-difference models.
Display table, graph, or spool output in the equation object window.
Display the contents of a table, graph, or spool in the window of the equation object.
Syntax
equation_name.display object_name
Examples
equation1.display tab1
Display the contents of the table TAB1 in the window of the object EQUATION1.
Cross-references
Most often used in constructing an EViews Add-in. See
“Custom Object Output”.
Display name for equation objects.
Attaches a display name to an equation which may be used to label output in place of the standard equation object name.
Syntax
equation_name.displayname display_name
Display names are case-sensitive, and may contain a variety of characters, such as spaces, that are not allowed in equation object names.
Examples
eq1.displayname Hours Worked
eq1.label
The first line attaches a display name “Hours Worked” to the equation EQ1, and the second line displays the label view of EQ1, including its display name.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Dynamic multipliers for long-run regressors in ARDL equations.
Displays a spool object with the cumulative dynamic multiplier curve for each of the long-run regressors. The argument is a positive integer denoting the horizon length, and defaults to 15.
Syntax
eq_name.dynmult(options) [horizon]
horizon is a positive integer denoting the horizon length, and defaults to 15.
Options
noci | Do not generate confidence intervals for asymmetric regressors. Note that confidence intervals can only be generated for asymmetric regressors. |
noshade | Display confidence interval using lines instead of shaded bands. |
level=number (default = 0.95) | Number between 0 and 1 representing the confidence interval level. |
reps=integer (default = 999) | Number of Monte Carlo repetitions used in the generation of confidence intervals (if applicable). |
f=number | Fraction of failed repetitions before stopping. Only applicable if a se_pattern is provided. |
prompt | Force the dialog to appear from within a program. |
p | Print output. |
Example
ardl_eq.dynmult
generates cumulative dynamic multiplier curves for each long-run regressor. The horizon length is 15, and the 95% confidence intervals (if they exist), are shaded, and derived from 999 Monte Carlo replications.
ardl_eq.dynmult(noshade) 30
generates cumulative dynamic multiplier curves for each long-run regressor. The horizon length is 30, and the 95% confidence intervals (if they exist), are not shaded.
ardl_eq.dynmult(noci)
produces cumulative dynamic multiplier curves for each long-run regressor. The horizon length is 15, and no confidence intervals are displayed.
ardl_eq.dynmult(level=0.99, reps=499) 10
shows cumulative dynamic multiplier curves for each long-run regressor. The horizon length is 10, and the 99% confidence intervals (if they exist), are shaded, and derived from 499 Monte Carlo replications.
Cross-references
See
“ARDL and Quantile ARDL” for a discussion of ARDL equation models.
Display a spool object showing tables with the conditional error correction (CEC) and error correction (EC) regression results in ARDL equations.
Syntax
eq_name.ecresults(options)
Options
Example
ardl_eq.ecresults
displays a spool object with the CEC and EC regressions from the ARDL equation ARDL_EQ.
Cross-references
See
“ARDL and Quantile ARDL” for a discussion of ARDL equation models.
Display the estimates of the fixed and/or random effects.
The effects view of a panel equation shows the estimates of the fixed and/or random effects associated with the estimated equation. These effects are expressed as deviations from the overall intercept displayed in the main equation output..
Syntax
eq_name.effects
Options
Examples
equation eq1.ls(cx=f) y c x1 x2
e1.effects
estimates the equation EQ1 with fixed effects, and displays a view showing the estimated cross-section deviations from the overall intercept.
Cross-references
See
“Panel Estimation” for a discussion of panel equation estimation.
Performs the regressor endogeneity test
The endogtest view of an equation carries out the Regressor Endogeneity/Donald-Wu Test for equations estimated via TSLS or GMM.
Syntax
eq_name.endogtest regressors
Options
prompt | Force the dialog to appear from within a program. |
Regressors
A list of equation regressors to be tested for endogeneity. Note the regressors must have been included in the original equation.
Examples
equation eq1.gmm y c x1 x2 @ z1 z2 z3 z4
e1.endogtest x1
estimates an equation, called EQ1, and estimates it via GMM, and then performs the Endogeneity Test, where X1 is tested for endogeneity.
Cross-references
Estimation of an elastic net model, including options for Lasso and ridge regression.
Syntax
equation_name.enet(options) y x1 [x2 x3 ...] [@vw(...)]
List the dependent variable first, followed by a list of the independent variables. Use a “C” if you wish to include an unpenalized intercept term.
Note that PDL and ARMA terms are not permitted in elastic net specifications.
If you wish to specify regressors with an individual penalty weight
, or to place inequality restrictions on the coefficient values, you may do so using special expressions of the form:
@vw(series_name, weight_value)
or
@vw(series_name[, wgt=weight_value, cmin=coef_min, cmax=coef_max])
where weight_value is a non-negative value, coef_min is a non-positive minimum coefficient value, and coef_max is a non-negative maximum coefficient value.
There are two forms of the special expression.
In the abbreviated form, you specify the variable name followed by the penalty weight value.
In the more general form, you specify the variable name followed by one or more keyword expressions in arbitrary order, with the “wgt=” argument specifying the penalty weight, “cmin=” with a non-positive minimum coefficient value, and “cmax=” with a non-negative maximum coefficient value.
When specifying individual regressor behavior using @vw, keep in mind that:
• The special intercept variable “C” is always non-penalized and has an implicit weight
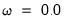
.
• Individual penalty weights may be also specified using a vector in the edit field on the dialog page (or using the command estimation option “lambdawgt=vector_name”). If the vector weights are specified and individual weights are specified using the @vw keyword, the vector weights will be applied first, followed by the individual variable weights.
• Individual coefficient limit values may also be specified using vectors in the and edit fields on the dialog page (or the command estimation options “coefmin=vector_name” and “coefmax=vector_name”). If vector coefficient limits are specified and individual regressor limits are specified using the @vw keyword, the vector limits will be applied first, followed by the individual limits weights.
EViews will normalize the individual penalty weights so that they sum to the number of coefficients.
Options
Specification Options
penalty=arg (default=“el”) | Type of threshold estimation: “enet” (elastic net), “ridge” (ridge), “lasso” (Lasso). |
alpha=arg (default=“.5”) | Value of the mixing parameter. Must be a value from zero to one. |
lambda=arg | Value(s) of the penalty parameter. Can be one or more numbers or vector objects.Values must be zero or greater. If left blank (default) EViews will generate a list. |
Penalty Options
ytrans=arg (default=“none”) | Scaling of the dependent variable: “none” (none), “L1” (L1), “L2” (L2), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
xtrans=arg (default=“stdpop”) | Scaling of the regressor variables: “none” (none), “L1” (L1 norm), “L2” (L2 norm), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
nlambdas=integer (default=100) | Number of penalty values for EViews-supplied list. |
lambdaratio=arg | Ratio of minimum to maximum lambda for EViews-supplied list. You may specify a value for the ratio parameter, or you may leave the edit field blank to let EViews specify a default value based on the number of observations 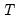 and the number of potential regressors 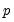 . By default, EViews will set the ratio to 0.001. |
lambdawgt= vector_name | Vector of individual penalty weights, containing non-negative values sized to and matching the order of the variables in the specification. If a vector is provided and individual weights are specified using one or more @vw regressors, the vector weights will be applied first, then overwritten by the individual variable weights. For comparability purposes, we normalize the final weights so that they sum to 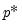 where 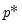 the number of non-zero  . |
nlambdamin=integer (default=5) | Minimum number of lambda values in the path before applying stopping rules. |
minddev=arg (default=1e-05) | Minimum change in deviance fraction to continue estimation. Truncate path estimation if relative change in deviance is smaller than this value. |
maxedev=arg (default=0.99) | Maximum of deviance explained fraction attained to terminate estimation. Truncate path estimation if fraction of null deviance explained is larger than this value. |
maxvars=arg | Maximum number of regressors in the model. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) reaches this value. |
maxvarsratio=arg | Maximum number of regressors in the model as a fraction of the number of observations. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) divided by the number of observations reaches this value. |
Cross Validation Options
cvmethod=arg (default=“kfold_cv”) | Cross-validation method: “kfold” (k-fold), “simple” (simple split), “mcarlo” (Monte Carlo), “leavepout” (leave-P-out), “leave1out” (leave-1-out), “rolling” (rolling window), “expanding” (expanding window). |
cvmeasure=arg (default=“mse”) | Cross-validation fit measure: “mse” (mean-squared error), “r2” (R‑squared), “mae” (mean absolute error), “mape” (mean absolute percentage error), “smape” (symmetric mean absolute percentage error). |
cvnfolds=arg (default=5) | Number of folds for K-fold cross-validation. For “cvmethod=kfold”. |
cvftrain=arg (default=0.8) | Proportion of data for split and Monte Carlo methods. For “cvmethod=simple” and “cvmethod=mcarlo”. |
cvnreps=arg (default=1) | Number of Monte Carlo method repetitions. For “cvmethod=mcarlo”. |
cvleaveout=arg (default=2) | Number of data points left out for leave-p-out method. For “cvmethod=leavepout”. |
cvnwindows=arg (default=4) | Number of windows for rolling window cross-validation method. For “cvmethod=rolling”. |
cvinitial=arg (default=12) | Number of initial data points in the training set for expanding cross-validation. For “cvmethod=expanding”. |
cvpregap=arg (default=0) | Number of observations between end of training set and beginning of test set. For “cvmethod=simple”, “cvmethod=rolling” and “cvmethod=expanding”. |
cvhorizon=arg (default=1) | Number of observation in the test set. For “cvmethod=rolling” and “cvmethod=expanding”. |
cvpostgap=arg (default=0) | Number of observations between end of test set and beginning of next training set for rolling window or between end of test set and end of next training set for expanding window. For “cvmethod=rolling” and “cvmethod=expanding” |
Random Number Options
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
rnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
Other Options
coefmin= vector_name, number | Vector of individual coefficient minimum values, containing negative or missing values sized to and matching the order of the variables in the specification, or a negative value for the minimum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual minimums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
coefmax= vector_name, number | Vector of individual coefficient maximum values, containing positive or missing values sized to and matching the order of the variables in the specification, or a positive value for the maximum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual maximums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
estmethod=arg | Estimation method: “cov” (use covariance algorithm) or “naive” (use naive algorithm). Default is EViews automatic. |
showopts / ‑showopts | [Do / do not] display estimation options in the output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic results view after estimation. |
Examples
The command
equation enet1.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
estimates an elastic net model with
equal to the default of 0.9, no regressor or dependent variable scaling, automatically determined 100-element lambda path with minimum lambda of 0.0001 times the maximum value, using the default K-fold cross-validation with 5 folds with an MSE objective and a random generator seed of 513255899 to determine the optimal value.
Similarly,
equation lasso1.enet(penalty=lasso, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
estimates a Lasso model with regressor population standard deviation scaling, with the remaining settings as before, while
equation ridge1.enet(penalty=ridge, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
estimates the equivalent ridge regression specification.
The command
equation enet2.enet(alpha=0.75, lambdaratio=.0001, cvmethod=rolling, cvmeasure=smape) lpsa c lcavol lweight age_s lbph svi lcp gleason pgg45
estimates an elastic net model with
equal to the 0.75 using rolling window cross-validation and SMAPE cross-validation.
We may use the @vw specifications to assign individual penalties and coefficient restrictions
equation enet3.enet(alpha=0.75, lambdaratio=.0001, cvmethod=rolling, cvseed=513255899) lpsa c @vw(lcavol, cmax=.4) lweight age lbph svi lcp gleason @vw(pgg45, cmax=0.075, w=1.2)
estimates an elastic net model with the coefficient of LCAVOL restricted to be less than or equal to 0.4, and the coefficient of PGG45 having a relative penalty weight of 1.2, and a maximum value of 0.075.
Identical specifications may be estimated using vectors of penalty weights and coefficient restrictions,
vector(9) cmax = na
cmax(2) = 0.4
cmax(9) = 1.2
vector(9) lwgt = 1
lwgt(9) = 1.2
equation enet4.enet(alpha=0.75, coefmax=cmax, lambdawgt=lwgt, lambdaratio=.0001, cvmethod=rolling, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason pgg45
and
vector(9) cmax = na
cmax(2) = 0.4
vector(9) lwgt = 1
lwgt(9) = 50
equation enet5.enet(alpha=0.75, coefmax=cmax, lambdawgt=lwgt, lambdaratio=.0001, cvmethod=rolling, cvseed=513255899) lpsa c lcavol lweight age lbph svi lcp gleason @vw(pgg45, cmax=0.075, w=1.2)
since the penalty weight for PGG45 in the vector is overwritten by the individual weight specified using the @vw.
Note that in neither case is the intercept penalized, even though the corresponding element of LWGT is equal to 1 since the specification of “C” is always implicitly treated as “@VW(C, 0)”.
Cross-references
See
“Elastic Net and Lasso” for a discussion of elastic net, ridge regression, and Lasso models.
Declare an equation object.
Syntax
equation eq_name
equation eq_name.method(options) specification
Follow the
equation keyword with a name and an optional specification. If you wish to enter the specification, you should follow the new equation name with a period, an estimation method, and the equation specification. Valid estimation methods are given in
“Equation Methods”. Refer to each method for a description of the available options.
Examples
equation cobdoug.ls log(y) c log(k) log(l)
declares and estimates an equation object named COBDOUG.
equation ces.ls log(y)=c(1)*log(k^c(2)+l^c(3))
declares an equation object named CES containing a nonlinear least squares specification.
equation demand.tsls q c p x @ x p(-1) gov
creates an equation object named DEMAND and estimates DEMAND using two-stage least squares with instruments X, lagged P, and GOV.
Cross-references
“Basic Regression Analysis” provides basic information on estimation and equation objects.
Factor breakpoint test for stability.
Carries out a factor breakpoint test for parameter constancy.
Syntax
eq_name.facbreak(options) ser1 [ser2 ser3 ...] @ x1 x2 x3
You must provide one or more series to be used as the factors with which to split the sample into categories. To specify more than one factor, separate the factors by a space. If the equation is specified by list and contains no nonlinear terms, you may specify a subset of the regressors to be tested for a breakpoint after an “@” sign.
Options
p | Print the result of the test. |
Examples
The commands:
equation ppp.ls log(spot) c log(p_us) log(p_uk)
ppp.facbreak season
perform a regression of the log of SPOT on a constant, the log of P_US, and the log of P_UK, and employ a factor breakpoint test to determine whether the parameters are stable through the different values of SEASON.
To test whether only the constant term and the coefficient on the log of P_US are “stable” enter the commands:
ppp.facbreak season @ c log(p_us)
Cross-references
See
“Factor Breakpoint Test” for further discussion.
Compute static forecasts or fitted values from an estimated equation.
When the regressor contains lagged dependent values or ARMA terms, fit uses the actual values of the dependent variable instead of the lagged fitted values. You may instruct fit to compare the forecasted data to actual data, and to compute forecast summary statistics.
Not available for equations estimated using ordered methods; use
Equation::makemodel to create a model using the ordered equation results (see example below).
Syntax
eq_name.fit(options) yhat [y_se]
eq_name.fit(options) yhat [y_se y_var]
Following the fit keyword, you should type a name for the forecast series and, optionally, a name for the series containing the standard errors. For ARCH specifications, you may use the second form of the command, and optionally include a name for the conditional variance series.
Forecast standard errors are currently not available for binary, censored, and count models.
Options
Basic Options
d | In models with implicit dependent variables, forecast the entire expression rather than the normalized variable. |
u | Substitute expressions for all auto-updating series in the equation. |
g | Graph the fitted values together with the ±2 standard error bands. |
ga | Graph the forecasts along with the actuals (if available). |
e | Produce the forecast evaluation table. |
i | Compute the fitted values of the index. Only for binary, censored and count models. |
s | Ignore ARMA terms and use only the structural part of the equation to compute the fitted values. |
n | Ignore coef uncertainty in standard error calculations that use them. |
forcsmpl = smpl | Fit sample (optional). If forecast sample is not provided, the workfile sample will be employed. |
f = arg (default= “actual”) | Out-of-fit-sample fill behavior: “actual” (fill observations outside the fit sample with actual values for the fitted variable), “na” (fill observations outside the fit sample with missing values). |
prompt | Force the dialog to appear from within a program. |
p | Print view. |
Stochastic Options
Options for forecasting from a functional coefficients estimated equation.
stochastic = arg (default = “none”) | Stochastic method: “none” (none), “mca” (Monte Carlo –asymptotic), “mcbs” (Monte Carlo – bootstrap), “bs” (bootstrap). |
reps = integer (default = 999) | Number of stochastic replications |
lhr = arg (default = 0.1) | Lower historical range (number between 0 and upper historical range). |
uhr = arg (default = 0.9) | Upper historical range (number between lower historical range and 1). |
bsdep | Bootstrap only the dependent variable (not the functional coefficient variable). |
Examples
equation eq1.ls cons c cons(-1) inc inc(-1)
eq1.fit c_hat c_se
genr c_up=c_hat+2*c_se
genr c_low=c_hat-2*c_se
line cons c_up c_low
The first line estimates a linear regression of CONS on a constant, CONS lagged once, INC, and INC lagged once. The second line stores the static forecasts and their standard errors as C_HAT and C_SE. The third and fourth lines compute the +/–2 standard error bounds. The fifth line plots the actual series together with the error bounds.
equation eq2.binary(d=l) y c wage edu
eq2.fit yf
eq2.fit(i) xbeta
genr yhat = 1-@clogit(-xbeta)
The first line estimates a logit specification for Y with a conditional mean that depends on a constant, WAGE, and EDU. The second line computes the fitted probabilities, and the third line computes the fitted values of the index. The fourth line computes the probabilities from the fitted index using the cumulative distribution function of the logistic distribution. Note that YF and YHAT should be identical.
Note that you cannot fit values from an ordered model. You must instead solve the values from a model. The following lines generate fitted probabilities from an ordered model:
equation eq3.ordered y c x z
eq3.makemodel(oprob1)
solve oprob1
The first line estimates an ordered probit of Y on a constant, X, and Z. The second line makes a model from the estimated equation with a name OPROB1. The third line solves the model and computes the fitted probabilities that each observation falls in each category.
Cross-references
To perform dynamic forecasting, use
Equation::forecast. See
Equation::makemodel and
Model::solve for forecasting from systems of equations or ordered equations.
See
“Forecasting from an Equation” for a discussion of forecasting in EViews and
“Discrete and Limited Dependent Variable Models” for a discussion of forecasting from binary, censored, truncated, and count models.
Detect outliers using the results of an estimated equation.
Use Tukey fences, mean/standard deviation fences, wavelet outliers, ARMA outliers or influence statistics to identify observations that may contain outliers.
Syntax
equation_name.resoutliers(options)
Options
sens=arg | Set the sensitivity level. Valid arguments are “low”, “medium” (default), and “high”. |
nofence | Do not perform Tukey and mean/standard deviation fences. |
nowave | Do not perform Wavelet Outlier detection. |
noarma | Do not perform ARMA based outlier detection. ARMA outlier detection is only available for least squares equations containing ARMA terms, and is turned on by default. |
noinf | Do not perform influence statistic (not including DFBETAS) based outlier detection. Influence statistic outlier detection is only available for linear least squares equations, and is turned on by default. |
dfbeta | Perform DFBETA influence statistic based outlier detection. DFBETA based outlier detection is only available for linear least squares equations, and is turned off by default. |
tukeyk=arg | Set the value k in the Tukey fence detection routine. This will override the value of k set by the sens= option. |
meanstdevk=arg | Set the value k in the mean/standard deviation fence detection routine. This will override the value of k set by the sens= option. |
wavesig=arg | Set the value false discovery rate significance value used in the Wavelet Outlier detection routine. This will override the value set by the sens= option. |
armac=arg | Set the value c in the ARMA outlier detection routine. This will override the value of c set by the sens= option. |
rsbound=arg | Set the value c in RSTUDENT outlier detection. This will override the value of c set by the sens= option. |
hbound=arg | Set the value c in HatMatrix outlier detection. This will override the value of c set by the sens= option. |
dfsbound=arg | Set the value c in DFFITS outlier detection. This will override the value of c set by the sens= option. |
covbound=arg | Set the value c in CovRatio outlier detection. This will override the value of c set by the sens= option. |
betabound=arg | Set the value c in DFBETA outlier detection. This will override the value of c set by the sens= option. |
series=name | Create a new series in the workfile, named name, containing a value of 1 for any observations identified as an outlier, and a value of 0 for any observation identified as not an outlier. |
datestring=name | Create a new string object in the workfile containing the dates (or observation identifiers) for any observations identified as an outlier. |
grlabels | Turn on observation labels on the outlier graph. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eq01.ls gdpc1 c unemp
eq01.resoutliers(nofence, dfbeta, sens=low)
Estimates an equation with GDPC1 as the dependent variable, and a constant and UNEMP as regressors. Then, outlier detection on the residuals is performed, opting to not use either fence detection, but to include the dfbeta influence statistics (along with the other influence statistics included by default), and setting the sensitivity of the detection to "low".
Cross-references
Test joint significance of the fixed effects estimates.
Tests the hypothesis that the estimated fixed effects are jointly significant using
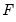
and LR test statistics. If the estimated specification involves two-way fixed effects, three separate tests will be performed; one for each set of effects, and one for the joint effects.
Syntax
eq_name.fixedtest(options)
Options
p | Print output from the test. |
Examples
equation eq1.ls(cx=f) sales c adver lsales
eq1.fixedtest
estimates a specification with cross-section fixed effects and tests whether the fixed effects are jointly significant.
Cross-references
See also
Equation::rcomptest for testing random for random components.
Computes dynamic forecasts of an estimated equation.
forecast computes the forecast for all observations in a specified sample. In some settings, you may instruct forecast to compare the forecasted data to actual data, and to compute summary statistics.
Syntax
eq_name.forecast(options) yhat [y_se]
eq_name.forecast(options) yhat [y_se y_var]
Enter a name for the forecast series and, optionally, a name for the series containing the standard errors. For ARCH specifications, you may use the second form of the command, and optionally enter a name for the conditional variance series. Forecast standard errors are currently not available for binary or censored models. forecast is not available for models estimated using ordered methods.
Options
d | In models with implicit dependent variables, forecast the entire expression rather than the normalized variable. |
u | Substitute expressions for all auto-updating series in the equation. |
g | Graph the forecasts together with the ±2 standard error bands. |
ga | Graph the forecasts along with the actuals (if available). |
e | Produce the forecast evaluation table. |
i | Compute the forecasts of the index. Only for binary, censored and count models. |
s | Ignore ARMA terms and use only the structural part of the equation to compute the forecasts. |
n | Ignore coef uncertainty in standard error calculations that use them. |
b =arg | MA backcast method: “fa” (forecast available). Only for equations estimated with MA terms. This option is ignored if you specify the “s” (structural forecast) option. The default method uses the estimation sample. |
forcsmpl=smpl | Forecast sample (optional). If forecast sample is not provided, the workfile sample will be employed |
f = arg (default= “actual”) | Out-of-forecast-sample fill behavior: “actual” (fill observations outside the forecast sample with actual values for the fitted variable), “na” (fill observations outside the forecast sample with missing values). |
stochastic | Perform stochastic simulation for dynamic equations estimated using least squares. |
streps=integer (default=1000) | Number of stochastic repetitions (for threshold regression or stochastic simulation). |
stfrac=number (default=.02) | Fraction of failed repetitions before stopping (for threshold regression or stochastic simulation). |
prompt | Force the dialog to appear from within a program. |
p | Print view. |
Examples
The following lines:
smpl 1970q1 1990q4
equation eq1.ls con c con(-1) inc
smpl 1991q1 1995q4
eq1.fit con_s
eq1.forecast con_d
plot con_s con_d
estimate a linear regression over the period 1970Q1–1990Q4, compute static (fitted) and dynamic forecasts for the period 1991Q1–1995Q4, and plot the two forecasts in a single graph.
equation eq1.ls m1 gdp ar(1) ma(1)
eq1.forecast m1_bj bj_se
eq1.forecast(s) m1_s s_se
plot bj_se s_se
estimates an ARMA(1,1) model, computes the forecasts and standard errors with and without the ARMA terms, and plots the two forecast standard errors.
Cross-references
To perform static forecasting with equation objects see
Equation::fit. For multiple equation forecasting, see
Equation::makemodel, and
Model::solve.
For more information on equation forecasting in EViews, see
“Forecasting from an Equation”.
Display functional coefficients bias results in graphical form.
For equations estimated using the functional coefficients method.
Syntax
eq_name.funbias(options)
Options
Basic Options
nolocalbw | Do not use the existing local pilot bandwidth (if it exists). Note that this option is only available if a local pilot bandwidth has been set previously, either through the estimation step, using the setpilotbw proc, or through another view or proc which set the local bandwidth. |
p | Print output. |
Pilot Bandwidth Options
If a local pilot bandwidth exists, all of the pilot bandwidth computation options below will be ignored unless the “nolocalbw” option is specified.
noupdate | Do not update the local pilot bandwidth using the current pilot bandwidth computation. |
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
auxk=integer (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage degree. This number should always be an even positive integer. |
Examples
eq1.funbias(p)
displays and prints a graph of the functional bias results for each functional coefficient computed using the existing local pilot bandwidth if present, or estimating the pilot bandwidth, if not. If estimated, the local pilot bandwidth is updated with the result.
eq1.funbias(nolocalbw, noupdate, plth=rsc)
ignores an existing local pilot bandwidth and instead computes a pilot bandwidth using the residual squares criterion. The existing saved local pilot bandwidth is retained.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation See
“Functional Bias” for discussion of bias.
Display functional coefficients final bandwidth selection results.
For equations estimated using the functional coefficients method.
Syntax
eq_name.funbw(options)
Options
Basic Options
pilot | Display pilot bandwidths. |
graph | Display results in graphical form. |
p | Print output. |
Examples
eq1.funbw
displays the bandwidth results in table form.
eq1.funbw(graph, p)
displays and prints a graph of the estimation final bandwidth selection results.
eq1.funbw(pilot)
displays a table for results for the estimation and local pilot bandwidths.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation. See
“Bandwidth Selection” and
“Bandwidth Views” for discussion of bandwidths.
Display graphs showing estimated lower and upper functional confidence intervals for the functional coefficients.
For equations estimated using the functional coefficients method.
Syntax
eq_name.funci(options) ci_level
where ci_level is a value between 0 and 1 specifying the confidence level you wish to display. If ci_level is omitted, EViews will use 0.95.
Options
Basic Options
nolocalbw | Do not use the existing local pilot bandwidth (if it exists). Note that this option is only available if a local pilot bandwidth has been set previously, either through the estimation step, using
Equation::setpilotbw, or through another view or proc which sets the local bandwidth. |
p | Print output. |
Confidence Interval Options
seband | Produce standard error bands instead of confidence intervals. |
sewidth =integer (default = 1) | Number of standard errors to use as half-width of confidence band (when “seband” is specified). |
nobias | Ignore bias in confidence interval determination. |
Pilot Bandwidth Options
If a local pilot bandwidth exists, all of the pilot bandwidth computation options below will be ignored unless the “nolocalbw” option is specified.
noupdate | Do not update the local pilot bandwidth using the current pilot bandwidth computation. |
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
auxk=integer (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage degree. This number should always be an even positive integer. |
Examples
eq1.funci 0.95 0.99
displays a graph of the 95% and 99% confidence intervals computed using the existing local pilot bandwidth if present, or estimating the pilot bandwidth, if not. If estimated, the local pilot bandwidth is updated with the result.
eq1.funci(seband, sewidth=3)
displays and prints a graph of the +/–3 confidence intervals.
The saved local pilot bandwidth is updated with the resulting bandwidth.
eq1.funci(nolocalbw, noupdate, plth=rsc) 0.9
ignores an existing local pilot bandwidth and instead computes a pilot bandwidth using the residual squares criterion and uses it in a 90% confidence interval calculation. The existing saved local pilot bandwidth is retained.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation. See
“Confidence Intervals” for discussion of functional confidence intervals.
Estimate a functional coefficient regression equation.
Syntax
eq_name.funcoef(options) y x1 [x2 x3 ...] @ funcoef_series
List the funcoef keyword, the dependent variable and a list of the regressor variables, followed by the “@” symbol and the name of the functional coefficient series.
Options
Basic Options
kern=arg (default=“epan”) | Kernel type: “epan” (Epanechnikov, default), “trngl” (Triangular), “unif” (Uniform), “gauss” (Normal–Gaussian), “bi” (Biweight–Quartic), “tri” (Triweight). |
eval=arg (default=“data”) | Evalution points: observed data (“data”), grid of values (“grid”). if “eval=grid” you must specify the grid values using “gmin=”, “gmax=” and “glen=”, or using “gvec=”. |
gmin = arg | Estimation grid minimum (if “eval=grid”). Must be specified along with “gmax=” and “glen=”. |
gmax = arg | Estimation grid maximum (if “eval=grid”). Must be specified along with “gmin=” and “glen=”. |
glen = arg | Estimation grid length (if “eval=grid”). Must be specified along with “gmin=” and “gmax=”. |
gvec = arg | Estimation grid points in a vector object (if “eval=grid”). |
plyk = arg (default = 1) | Estimation polynomial degree for final stage. |
auxk = arg (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage. |
p | Print results. |
Pilot Bandwidth Options
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
Final Bandwidth Options
fnlh =arg (default = “cv”) | Final bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), integrated asymptotic mean square error (“mse”), leave-one-out cross-validation (“loo”), nonparametric AIC (“aic”), user-defined (“user”). |
fnlbw=arg | User-defined bandwidth (if “fnlh=user”). |
fnlhmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “fnlh=user”). |
fnlhmax=arg (default =1) | Bandwidth grid search maximum value (if not “fnlh=user”). |
fnlhlen=integer (default = 100) | Bandwidth grid search length (if not “fnlh=user”). |
fnlhinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “fnlh=user”). |
fnlhcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (if not “fnlh=user”). |
fnlm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “fnlh=cv”). |
fnlfq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “fnlh=cv”). |
Examples
We consider examples for three equations that estimate FCOEF using UNRATE as the dependent variable, UNRATE(-1 to -2) as independent variables, and LWAGE(-4) as the functional coefficient variable.
eq.funcoef(eval=grid, gmin=0, gmax=10, glen=100) unrate unrate(-1) unrate(-2) @ lwage(-4)
evaluates over a custom uniform grid from 0 to 10 with length 100.
eq.funcoef(eval=grid, gvec=vecgrid) unrate unrate(-1) unrate(-2) @ lwage(-4)
evaluates over a custom grid provided by the values of a workfile vector called VECGRID.
eq.funcoef(plyk=3, auxk=5) unrate unrate(-1) unrate(-2) @ lwage(-4)
estimates using local polynomial fitting with main polynomial degree 3 and auxiliary polynomial degree 5. The latter is employed deriving bias, variance, and bandwidths.
Cross-references
See
“Functional Coefficient Regression” for additional discussion of functional coefficients estimation.
Display functional coefficient covariance functions in graphical form.
For equations estimated using the functional coefficients method.
Syntax
eq_name.funcov(options)
Options
Basic Options
corr | Produce correlations instead of covariances. |
nolocalbw | Do not use the existing local pilot bandwidth (if it exists) Note that this option is only available if a local pilot bandwidth has been set previously, either through the estimation step, using
Equation::setpilotbw, or through another view or proc which sets the local bandwidth. |
p | Print output. |
Pilot Bandwidth Options
If a local pilot bandwidth has exists, all of the pilot bandwidth computation options below will be ignored unless the “nolocalbw” option is specified.
noupdate | Do not update the local pilot bandwidth using the current pilot bandwidth computation. |
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
auxk=integer (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage degree. This number should always be an even positive integer. |
Examples
eq1.funcov
displays the functional covariances computed using the existing local pilot bandwidth if present, or estimating the pilot bandwidth, if not. If estimated, the local pilot bandwidth is updated with the result.
eq1.funcov(corr, p)
displays and prints a graph of the correlation results.
eq1.funcov(nolocalbw, noupdate, auxk=4, plth=rsc)
ignores an existing local pilot bandwidth and instead computes a pilot bandwidth using the residual squares criterion and auxiliary degree of 4, and uses it in the covariance calculation. The existing saved local pilot bandwidth is retained.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation. See
“Functional Covariance/Correlation Matrix” for discussion of functional covariances.
Perform functional coefficients hypothesis and stability tests.
Test one or more functional coefficients for stability or equality with values in a specified series.
For equations estimated using the functional coefficients method.
Syntax
eq_name.funtest(options) arg
where arg is a comma separated list of functional coefficient restrictions for which you wish separately to test.
The coefficient restrictions
coefspec = restriction
where coefspec is a coefficient spec consisting of,
• c(k)
•
for individual coefficient k, and all coefficients, respectively, and restriction takes the form:
• series_name or vector
• scalar
•
Options
Basic Options
nolocalbw | Do not use the existing local pilot bandwidth (if it exists) Note that this option is only available if a local pilot bandwidth has been set previously, either through the estimation step, using
Equation::setpilotbw, or through another view or proc which sets the local bandwidth. |
p | Print output. |
Pilot Bandwidth Options
If a local pilot bandwidth has exists, all of the pilot bandwidth computation options below will be ignored unless the “nolocalbw” option is specified.
noupdate | Do not update the local pilot bandwidth using the current pilot bandwidth computation. |
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
auxk=integer (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage degree. This number should always be an even positive integer. |
Examples
eq1.funtest @all=10, c(2)=2, c(3)=y
performs three separate tests for the restrictions: all of the coefficients are constant and equal to 10, c(2) is a constant equal to 2, and c(3) is equal to the series Y using an existing local pilot bandwidth if present, or estimating the pilot bandwidth, if not. If estimated, the local pilot bandwidth is updated with the result.
eq1.funtest(nolocalbw, noupdate, auxk=4, plth=rsc) @all=3, c(2)=2, c(3)=y
ignores an existing local pilot bandwidth and instead computes a pilot bandwidth using the residual squares criterion and auxiliary degree of 4, and uses it in the test calculation. The existing saved local pilot bandwidth is retained.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation. See
“Stability and Significance Tests” for discussion of functional covariances.
Conditional variance/covariance of (G)ARCH estimation.
Displays the conditional variance, covariance or correlation of an equation estimated by ARCH.
Syntax
eq_name.garch(options)
Options
v | Display conditional variance graph instead of the standard deviation graph. |
p | Print the graph |
Examples
equation eq1.arch sp500 c
eq1.garch
estimates a GARCH(1,1) model and displays the estimated conditional standard deviation graph.
eq1.garch(v, p)
displays and prints the estimated conditional variance graph.
Cross-references
ARCH estimation is described in
“ARCH and GARCH Estimation”.
Estimate a Generalized Linear Model (GLM).
Syntax
eq_name.glm(options) spec
List the glm keyword, followed by the dependent variable and a list of the explanatory variables, or an explicit linear expression.
If you enter an explicit linear specification such as “Y=C(1)+C(2)*X”, the response variable will be taken to be the variable on the left-hand side of the equality (“Y”) and the linear predictor will be taken from the right-hand side of the expression (“C(1)+C(2)*X”).
Offsets may be entered directly in an explicit linear expression or they may be entered as using the “offset=” option.
Specification Options
family=arg (default=“normal”) | Distribution family: Normal (“normal”), Poisson (“poisson”), Binomial Count (“binomial”), Binomial Proportion (“binprop”), Negative Binomial (“negbin”), Gamma (“gamma”), Inverse Gaussian (“igauss”), Exponential Mean (“emean)”, Power Mean (“pmean”), Binomial Squared (“binsq”). The Binomial Count, Binomial Proportion, Negative Binomial, and Power Mean families all require specification of a distribution parameter: |
n=arg (default=1) | Number of trials for Binomial Count (“family=binomial”) or Binomial Proportions (“family=binprop”) families. |
fparam=arg | Family parameter value for Negative Binomial (“family=negbin”) and Power Mean (“family=pmean”) families. |
link=arg (default=“identity”) | Link function: Identity (“identity”), Log (“log”), Log Compliment (“logc”), Logit (“logit”), Probit (“probit”), Log-log (“loglog”), Complementary Log-log (“cloglog”), Reciprocal (“recip”), Power (“power”), Box-Cox (“boxcox”), Power Odds Ratio (“opow”), Box-Cox Odds Ratio (“obox”). The Power, Box-Cox, Power Odds Ratio, and Box-Cox Odds Ratio links all require specification of a link parameter specified using “lparam=”. |
lparam=arg | Link parameter for Power (“link=power”), Box-Cox (“link=boxcox”), Power Odds Ratio (“link=opow”) and Box-Cox Odds Ratio (“link=obox”) link functions. |
offset=arg | Offset terms. |
disp=arg | Dispersion estimator: Pearson 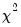 statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”). The default is family specific: “unit” for Binomial Count, Binomial Proportion, Negative Binomial, and Poison, and “pearson” for all others. The “deviance” option is only offered for families in the exponential family of distributions (Normal, Poisson, Binomial Count, Binomial Proportion, Negative Binomial, Gamma, Inverse Gaussian). |
dispval=arg | User-dispersion value (if “disp=user”). |
fwgts=arg | Frequency weights. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
In addition to the specification options, there are options for estimation and covariance calculation.
Additional Options
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “fisher” (IRLS – Fisher Scoring), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
estmeth=arg (default=”marquardt”) | Legacy estimation algorithm: Quadratic Hill Climbing (“marquardt”), Newton-Raphson (“newton”), IRLS - Fisher Scoring (“irls”), BHHH (“bhhh”). (Applicable when “optmethod=legacy”.) |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
preiter=arg (default=0) | Number of IRLS pre-iterations to refine starting values (only available for non-IRLS estimation). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method), “glm” (GLM method), “cr” (cluster robust). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian), “fisher” (expected Hessian). (Applicable when “optmethod=” not equal to “legacy”. |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate.(For non-cluster robust methods). |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). Applicable where “cov=hac”. |
covinfosel=arg (default=”aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). For settings where “cov=hac, covlag=a”. |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of  . For settings where “cov=hac, covlag=a”. |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). For settings where “cov=hac”. |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). For settings where “cov=hac” and “covkern=” is specified. |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). For settings where “cov=hac” and “covkern=” is specified. |
covbwoffset=number | Apply offset to automatically selected bandwidth. For settings where “cov=hac”, “covkern=” is specified, and “covbw=” is not user-specified. |
covbwint | Use integer portion of kernel bandwidth. For settings where “cov=hac” and “covkern=” is specified. |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eqstrike.glm(link=log) numb c ip feb
estimates a normal regression model with exponential mean.
equation eqbinom.glm(family=binomial, n=total) disease c snore
estimates a binomial count model with default logit link where TOTAL contains the number of binomial trials and DISEASE is the number of binomial successes. The specification
equation eqbinom.glm(family=binprop, n=total, cov=huber, nodf) disease/total c snore
estimates the same specification in proportion form, and computes the coefficient covariance using the Huber-White sandwich with no d.f. correction.
equation eqprate.glm(family=binprop, disp=pearson) prate mprate log(totemp) log(totemp)^2 age age^2 sole
estimates a binomial proportions model with default logit link, but computes the coefficient covariance using the GLM scaled covariance with dispersion computed using the Pearson Chi-square statistic.
equation eqprate.glm(family=binprop, link=probit, cov=huber) prate mprate log(totemp) log(totemp)^2 age age^2 sole
estimates the same basic specification, but with a probit link and Huber-White standard errors.
equation testeq.glm(family=poisson, offset=log(pyears)) los hmo white type2 type3 c
estimates a Poisson specification with an offset term LOG(PYEARS).
Cross-references
Estimation by generalized method of moments (GMM).
The equation object must be specified with a list of instruments.
Syntax
eq_name.gmm(options) y x1 [x2 x3...] @ z1 [z2 z3...]
eq_name.gmm(options) specification @ z1 [z2 z3...]
Follow the name of the dependent variable by a list of regressors, followed by the “@” symbol, and a list of instrumental variables which are orthogonal to the residuals. Alternatively, you can specify an expression using coefficients, an “@” symbol, and a list of instrumental variables. There must be at least as many instrumental variables as there are coefficients to be estimated.
In panel settings, you may specify dynamic instruments corresponding to predetermined variables. To specify a dynamic instrument, you should tag the instrument using “@DYN”, as in “@DYN(X)”. By default, EViews will use a set of period-specific instruments corresponding to lags from -2 to “-infinity”. You may also specify a restricted lag range using arguments in the “@DYN” tag. For example, to use lags from-5 to “-infinity” you may enter “@DYN(X, -5)”; to specify lags from -2 to -6, use “@DYN(X, -2, -6)” or “@DYN(X, -6, -2)”.
Note that dynamic instrument specifications may easily generate excessively large numbers of instruments.
Options
Non-Panel GMM Options
Basic GMM Options
nocinst | Do not include automatically a constant as an instrument. |
method=keyword | Set the weight updating method. keyword should be one of the following: “nstep” (N-Step Iterative, or Sequential N-Step Iterative, default), “converge” (Iterate to Convergence or Sequential Iterate to Convergence), “simul” (Simultaneous Iterate to Convergence), “oneplusone” (One-Step Weights Plus One Iteration), or “cue” (Continuously Updating”. |
gmmiter=integer | Number of weight iterations. Only applicable if the “method=nstep” option is set. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
m=integer | Maximum number of iterations. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list (see also
param). |
s=number | Determine starting values for equations specified by list. Specify a number between zero and one representing the fraction of preliminary TSLS estimates computed without AR or MA terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR and MA terms which are instead set to EViews determined default values. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get the one-step weighting matrix. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Estimation Weighting Matrix Options
instwgt=keyword | Set the estimation weighting matrix type. Keyword should be one of the following: “tsls” (two-stage least squares), “white” (White diagonal matrix), “hac” (Newey-West HAC, default) or “user” (user defined). |
instwgtmat=name | Set the name of the user-defined estimation weighting matrix. Only applicable if the “instwgt=user” option is set. |
instlag=arg (default=1) | Whitening Lag specification: integer (user-specified lag value), “a” (automatic selection). |
instinfosel=arg (default=“aic”) | Information criterion for automatic whitening lag selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “instlag=a”). |
instmaxlag= integer | Maximum lag-length for automatic selection (optional) (if “instlag=a”). The default is an observation-based maximum of  . |
instkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
instbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
instnwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “instbw=neweywest”). |
instbwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
instbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Covariance Options
cov=keyword | Covariance weighting matrix type (optional): “updated” (estimation updated), “tsls” (two-stage least squares), “white” (White diagonal matrix), “hac” (Newey-West HAC), “wind” (Windmeijer) “cr” (cluster robust). or “user” (user defined). The default is to use the estimation weighting matrix. |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. (For non-cluster robust methods). |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=”aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 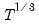 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
covbwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
Panel GMM Options
cx=arg | Cross-section effects method: (default) none, fixed effects estimation (“cx=f”), first-difference estimation (“cx=fd”), orthogonal deviation estimation (“cx=od”) |
per=arg | Period effects method: (default) none, fixed effects estimation (“per=f”). |
levelper | Period dummies always specified in levels (even if one of the transformation methods is used, “cx=fd” or “cx=od”). |
wgt=arg | GLS weighting: (default) none, cross-section system weights (“wgt=cxsur”), period system weights (“wgt=persur”), cross-section diagonal weighs (“wgt=cxdiag”), period diagonal weights (“wgt=perdiag”). |
gmm=arg | GMM weighting: 2SLS (“gmm=2sls”), White period system covariances (Arellano-Bond 2-step/n-step) (“gmm=perwhite”), White cross-section system (“gmm=cxwhite”), White diagonal (“gmm=stackedwhite”), Period system (“gmm=persur”), Cross-section system (“gmm=cxsur”), Period heteroskedastic (“cov=perdiag”), Cross-section heteroskedastic (“gmm=cxdiag”). By default, uses the identity matrix unless estimated with first difference transformation (“cx=fd”), in which case, uses (Arellano-Bond 1-step) difference weighting matrix. In this latter case, you should specify 2SLS weights (“gmm=2sls”) for Anderson-Hsiao estimation. |
cov=arg | Coefficient covariance method: (default) ordinary, White cross-section system robust (“cov=cxwhite”), White period system robust (“cov=perwhite”), White heteroskedasticity robust (“cov=stackedwhite”), Cross-section system robust/PCSE (“cov=cxsur”), Period system robust/PCSE (“cov=persur”), Cross-section heteroskedasticity robust/PCSE (“cov=cxdiag”), Period heteroskedasticity robust (“cov=perdiag”). |
keepwgts | Keep full set of GLS/GMM weights used in estimation with object, if applicable (by default, only weights which take up little memory are saved). |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
iter=arg (default=“onec”) | Iteration control for GLS and GMM weighting specifications: perform one weight iteration, then iterate coefficients to convergence (“iter=onec”), iterate weights and coefficients simultaneously to convergence (“iter=sim”), iterate weights and coefficients sequentially to convergence (“iter=seq”), perform one weight iteration, then one coefficient step (“iter=oneb”). |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list (see also
param). |
s=number | Determine starting values for equations specified by list. Specify a number between zero and one representing the fraction of preliminary TSLS estimates computed without AR terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR terms which are instead set to EViews determined default values. |
m=integer | Maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
l=number | Set maximum number of iterations on the first-stage iteration to get the one-step weighting matrix. |
unbalsur | Compute SUR factorization in unbalanced data using the subset of available observations for a cluster. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Note that some options are only available for a subset of specifications.
Examples
In a non-panel workfile, we may estimate equations using the standard GMM options. The specification:
gmmc.gmm(instwgt=white,gmmiter=2,nodf) cons c y y(-1) w @ c p(-1) k(-1) x(-1) tm wg g t
estimates the Klein equation for consumption using GMM with a White diagonal weighting matrix (two steps and no degree of freedom correction). The command:
gmmi.gmm(method=cue,instwgt=hac,instlag=1,instkern=thann,instbw=andrews,nodf) i c y y(-1) k(-1) @ c p(-1) k(-1) x(-1) tm wg g t
estimates the Klein equation for investment using a Newey-West HAC weighting matrix, with pre-whitening with 1 lag, a Tukey-Hanning kernel and the Andrews automatic bandwidth routine. The estimation is performed using continuously updating weight iterations.
When working with a workfile that has a panel structure, you may use the panel equation estimation options. The command
eq.gmm(cx=fd, per=f) dj dj(-1) @ @dyn(dj)
estimates an Arellano-Bond “1-step” estimator with differencing of the dependent variable DJ, period fixed effects, and dynamic instruments constructed using DJ with observation specific lags from period
to 1.
To perform the “2-step” version of this estimator, you may use:
eq.gmm(cx=fd, per=f, gmm=perwhite, iter=oneb) dj dj(-1) @ @dyn(dj)
where the combination of the options “gmm=perwhite” and (the default) “iter=oneb” instructs EViews to estimate the model with the difference weights, to use the estimates to form period covariance GMM weights, and then re-estimate the model.
You may iterate the GMM weights to convergence using:
eq.gmm(cx=fd, per=f, gmm=perwhite, iter=seq) dj dj(-1) @ @dyn(dj)
Alternately:
eq.gmm(cx=od, gmm=perwhite, iter=oneb) dj dj(-1) x y @ @dyn(dj,-2,-6) x(-1) y(-1)
estimates an Arellano-Bond “2-step” equation using orthogonal deviations of the dependent variable, dynamic instruments constructed from DJ from period
to
, and ordinary instruments X(-1) and Y(-1).
Cross-references
See
“Generalized Method of Moments” and
“Panel Estimation” for discussion of the various GMM estimation techniques.
Gradients of the objective function.
Displays the gradients of the objective function. Evaluating the gradients at current coefficient values allows you to examine the behavior of the objective function at starting values.
The (default) summary form shows the value of the gradient vector at the estimated parameter values (if valid estimates exist) or at the current coefficient values.
You may optionally choose to display the results in tabular or graphical form. The tabular form shows a spreadsheet view of the gradients for each observation. The graphical form shows this information in a multiple line graph.
Syntax
equation_name.grads(options)
Options
t | Display spreadsheet view of the values of the gradients of the objective function with respect to the coefficients evaluated at each observation. |
g | Display multiple graph showing the gradients of the objective function with respect to the coefficients evaluated at each observation. |
p | Print results. |
Examples
To show a summary view of the gradients:
eq1.grads
To display and print the table view:
eq1.grads(t, p)
Cross-references
Estimate a selection equation using the Heckman ML or 2-step method.
Syntax
equation_name.heckit(options) response_eqn @ selection_eqn
The response equation should be the dependent variable followed by a list of regressors. The selection equation should be a binary dependent variable followed by a list of regressors.
Options
General Options
2step | Use the Heckman 2-step estimation method. Note that this option is incompatible with the maximum likelihood options below. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print the estimation results. |
ML Options
Note these options are not available if the "2step" option, above, is used.
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich methods)., |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
m=integer | Set maximum number of iterations. |
c=number | Set convergence criteria. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
s=number | Scale EViews’ starting values by number. |
r | Use Newton-Raphson optimizer. |
b | Use BHHH optimizer. |
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-1.txt
equation eq01.heckit ww c ax ax^2 we cit @ lfp c wa wa^2 faminc we (k618+kl6)>0
equation eq02.heckit(2step) ww c ax ax^2 we cit @ lfp c wa wa^2 faminc we (k618+kl6)>0
This example replicates the Heckman Selection example given in Greene (2008, page 888), which uses data from the Mroz (1987) study to estimate a selection model. The first line of this example downloads the data set, the second line creates an equation object and estimates it using the default maximum likelihood estimation method of Heckman Selection, which replicates the first pane of Table 24.3 in Greene. The third line estimates the same model, using the two-step approach, which replicates the second pane of Table 24.3.
Cross-references
Test for Heteroskedasticity.
Performs a test for heteroskedasticity among the residuals from an equation.
The test performed can be a Breusch-Pagan-Godfrey (the default option), Harvey, Glejser, ARCH or White style test.
Syntax
equation_name.hettest(options) variables
Options
type = keyword | where keyword is either “BPG” (Breusch-Pagan-Godfrey - default), “Harvey”, “Glejser”, “ARCH”, or “White”. |
c | include cross terms for White test. |
lags = int | set number of lags to use for ARCH test. (Only applies when type = “ARCH”. |
prompt | Force the dialog to appear from within a program. |
Variables
A list of series names to be included in the auxiliary regression. Not applicable for ARCH or White type tests. The following keywords may be included:
@regs | include every regressor from the original equation. |
@grads | include every gradient in the original equation (non-linear equations only). |
@grad(int) | include the int-th gradient. |
@white(key) | include white-style regressors (the cross-product of the regressor list, or the gradient list if non-linear). key may be on of the following keywords: “@regs” (include every regressor from the original equation), “@drop(variables)” (drop a variable from those already included. For example, “@white(@regs @drop(x2))” would include all original regressors apart from X2), “@comp” (include the compatible style White regressors, i.e. levels, squares, and cross-products). |
@arch(lag_structure) | include an ARCH specification with the number of lags specified by lag_structure. If lag_structure is a single number, then it defines the number of lags to include. Otherwise, the lag structure is in pairs. For example, “@arch(1 5 9 10)” will include lags 1, 2, 3, 4, 5, 9, 10. |
@uw(variables) | include unweighted variables (only applicable in a weighted original equation). |
Examples
eq1.hettest(type=harvey) @white(@regs @drop(log(ip)))
performs a heteroskedasticity test with an auxiliary regression of the log of squared residuals on the cross product of all the original equation’s variables, except LOG(IP).
Cross-references
See
“Heteroskedasticity Tests” for a discussion of heteroskedasticity testing in EViews.
Histogram and descriptive statistics of the residual series of an equation.
Syntax
equation_name.hist(options)
Options
Examples
eq1.hist
Displays the histogram and descriptive statistics of the residual series of equation EQ1.
Cross-references
See
“Histogram and Stats” for a discussion of the descriptive statistics reported in the histogram view.
Display a graph of the selection criteria for the top 20 models observed as part of model selection during estimation.
This view is only available for equations estimated using the ARDL or TAR methods.
Syntax
equation_name.icgraph
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
equation eq01.ardl(deplags=8, reglags=8) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
show eq01.icgraph
This example uses data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000. The first line of this example downloads the data set, the second line creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable, and the log of real GDP as a dynamic regressor. Quarterly dummy variables are included as static regressors. Automatic model selection is used.
The final line of code displays a graph showing the Akaike information criteria (the default selection method) for each of the models estimates.
Cross-references
Display a table of the log-likelihood and selection criteria for the top 20 models observed as part of model selection during estimation.
This view is only available for equations estimated using the ARDL or TAR methods.
Syntax
equation_name.ictable
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
equation eq01.ardl(deplags=8, reglags=8) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
show eq01.ictable
This example uses data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000. The first line of this example downloads the data set, the second line creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable, and the log of real GDP as a dynamic regressor. Quarterly dummy variables are included as static regressors. Automatic model selection is used.
The final line of code displays a table showing the log-likelihood value, Akaike information criteria, Schwarz information criteria, the Hannan-Quinn criteria and the adjusted R-squared of the top 20 models.
Cross-references
Scaled difference in the estimated betas for influence statistics.
DFBETAS are the scaled difference in the estimated betas between the original equation and an equation estimated without that observation.
Syntax
equation_name.infbetas(options) [base_name]
where base_name is an optional naming suffix used to store the DFBETAS into the workfile.
Options
t | Show a table of the statistics (the default is to display a graph view of the specified statistics). |
rows = key | The number of observations/rows to display in the table, where key can be either “50”, “100” (default), “150”, or “200”. |
g=arg | arg is the name of an object in which the graph output will be saved. |
prompt | Force the dialog to appear from within a program. |
Examples
equation eq1.ls y c x z
eq1.infbetas
displays a graph of the DFBETAS corresponding to the coefficients for C, X, and Z.
eq1.infbetas(t) out
will display a table showing the first 150 rows of DFBETAs in table form and saves the results in the series COUT, XOUT and ZOUT.
Cross-references
Influence statistics.
Displays influence statistics to discover influential observations, or outliers.
Syntax
equation_name.infstats(options)
equation_name.infstats(options) stats_list [@ save_names]
If no stats_list is provided all of the statistics will be displayed. save_names is an optional list of names for storing the statistics into series in the workfile. The save_names should match the order in which the keywords in stats_list are entered.
Options
t | Show a table of the statistics (the default is to display a graph view of the specified statistics). |
rows = key | The number of observations/rows to display in the table, where key can be either “50”, “100” (default), “150”, or “200”. |
sort = key | Sort order for the table, where key can be “r” (Residual - default), “rs” (RStudent), “df” (DFFITS), “dr” (Dropped Residual), “cov” (COVRATIO), “h” (diagonal elements of the hat matrix). |
sortdisp | Display the table by the sort order rather than by the observation order. |
prompt | Force the dialog to appear from within a program. |
The stats_list parameter is a list of keywords indicating which statistics to display. It may take on the values:
rstudent | The studentized residual: the t-statistic on a dummy variable that is equal to 1 on that observation only. |
dffits | The scaled difference in fitted values. |
drresid | Dropped residual: the estimated residual for that observation had the equation been run without that observation. |
covratio | The ratio of the covariance matrix of the coefficients with and without that observation. |
hatmatrix | Diagonal elements of the hat matrix:  |
Examples
eq1.infstats(t, rows=150, sort=rs) rstudent covratio dffits @ rstuds covs
will display a table showing the 150 largest RSTUDENT statistics, along with the corresponding COVRATIO and DFFITS statistics. It will save the RSTUDENT and COVRATIO statistics into the series in the workfile named RSTUDS and COVS, respectively.
Cross-references
Shows a summary of the equation instruments.
Changes the view of the equation to the Instrument Summary view. Note this is only available for equations estimated by TSLS, GMM, or LIML.
Syntax
eq_name.instsum
Examples
equation eq1.tsls sales c adver lsales @ gdp unemp int
e1.instsum
creates an equation E1 and estimates it via two-stage least squares, then shows a summary of the instruments used in estimation.
Cross-references
See
“Instrument Summary” for discussion.
Display or change the label view of an equation, including the last modified date and display name (if any).
As a procedure, label changes the fields in the equation label.
Syntax
equation_name.label
equation_name.label(options) [text]
Options
The first version of the command displays the label view of the equation. The second version may be used to modify the label. Specify one of the following options along with optional text. If there is no text provided, the specified field will be cleared.
c | Clears all text fields in the label. |
d | Sets the description field to text. |
s | Sets the source field to text. |
u | Sets the units field to text. |
r | Appends text to the remarks field as an additional line. |
p | Print the label view. |
Examples
The following lines replace the remarks field of EQ1 with “Data from CPS 1988 March File”:
eq1.label(r)
eq1.label(r) Data from CPS 1988 March File
To append additional remarks to EQ1, and then to print the label view:
eq1.label(r) Log of hourly wage
eq1.label(p)
To clear and then set the units field, use:
eq1.label(u) Millions of bushels
Cross-references
See
“Labeling Objects” for a discussion of labels.
Display the spreadsheet of the matrix of coefficient values along the lambda path.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
Rows will contain coefficients for a given lambda; columns contain coefficients for specific variables.
The row displaying the model selected optimal lambda will be highlighted.
Only coefficients that have non-zero values for at least one lambda in the path will be displayed.
Syntax
eq_name.lambdacoefs(options)
Options
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.lambdacoefs
displays a table of the anywhere non-zero coefficients, with rows corresponding to values of lambda in the path, and columns corresponding to different variables. The row with the cross-validation selected optimal value of lambda will be highlighted.
Cross-references
For further discussion, see
“Elastic Net and Lasso”.
The data underlying this table are available via the data member @lambdacoefs.
Display the table showing various values associated with estimation along the lambda path.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
Display a table showing the number of non-zero coefficients, estimation objective, sums-of-squared residuals penalty, and where applicable, the L1-norm and the L2-norm penalty portions of the objective associated with each lambda in the path.
The row displaying the model selected optimal lambda will be highlighted.
Syntax
eq_name.lambdaest(options)
Options
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.lambdaest
displays the table showing various estimation statistics associated with each lambda in the path.
Cross-references
For further discussion, see
“Elastic Net and Lasso”.
The data underlying this table are available via the data member @lambdaest.
Display the table showing various fit statistics associated with estimates along the lambda path.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
Display a table showing the number of non-zero coefficients, R-squared, adjusted R-squared, standard error of the regression, and sums-of-squared residuals associated with each of the lambda in the path.
The row displaying the model selected optimal lambda will be highlighted.
Syntax
eq_name.lambdafit(options)
Options
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.lambdafit
displays the table showing various fit statistics associated with each lambda in the path.
Cross-references
For further discussion, see
“Elastic Net and Lasso”.
The data underlying this table are available via the data member @lambdafit.
Display graphs of lambda against various fit and estimation measures.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
You may plot lambda against the paths of: the number of non-zero coefficients, model selection objective, R-squared and adjusted R-square fit statistics, standard error of the regression, sum-of-squared residuals, L1-norm coefficient penalty, L2-norm squared coefficient penalty, and the estimation objective.
A vertical line will be included to identify the selected optimal lambda.
By default, EViews will display a spool object containing all of the plots. You may use the “type=” option to produce a specific graph.
Syntax
eq_name.lambdapaths(options)
Options
type=arg | Graph of the log lambda against the path of: “nonzero” (number of non-zero coefficients), “model” (model selection objective), “fit” (R-squared and adjusted R-squared fit statistics), “se” (standard error of regression), “ssr” (sum-of-squared residuals), “l1” (L1 coefficient penalty, if applicable), “l2” (L2-squared coefficient penalty, if applicable), “estobj” (estimation objective). If “type=” is not provided, EViews will display the spool object all of the graphs. |
p | Print output. |
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.lambdapath
displays estimates a spool containing graphs of log lambda plotted against paths of the number of non-zero coefficients, fit measures, and estimation measures.
my_eq.lambapath(type=nonzero, p)
displays and prints a single graph of log lambda plotted against the number of non-zero coefficients, while
my_eq.lambapath(type=fit)
plots log lambda against the paths of the R-squared and adjusted R-squared.
Cross-references
For further discussion, see
“Elastic Net and Lasso”The data underlying these graphs are available via the data members @lambdafit, and @lambdaest.
Limited Information Maximum Likelihood and K-class Estimation.
Syntax
eq_name.liml(options) y c x1 [x2 x3 ...] @ z1 [z2 z3 ...]
eq_name.liml(options) specification @ z1 [z2 z3 ...]
To use the liml command, list the dependent variable first, followed by the regressors, then any AR or MA error specifications, then an “@”-sign, and finally, a list of exogenous instruments.
You may estimate nonlinear equations or equations specified with formulas by first providing a specification, then listing the instrumental variables after an “@”-sign. There must be at least as many instrumental variables as there are independent variables. All exogenous variables included in the regressor list should also be included in the instrument list. A constant is included in the list of instrumental variables, unless the noconst option is specified.
Options
noconst | Do not include a constant in the instrumental list. Without this option, a constant will always be included as an instrument, even if not specified explicitly. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
kclass=number | Set the value of 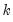 in the K‑class estimator. If omitted, LIML is performed, and 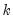 is calculated as part of the estimation procedure. |
se = arg (default=“iv”) | Set the standard-error calculation type: IV based (“se=iv”), K-Class based (“se=kclass”), Bekker (“se=bekk”), or Hansen, Hausman, and Newey (“se=hhn”). |
m=integer | Set maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. Available only for legacy estimation (“optmeth=legacy”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
equation eq1.liml gdp c cpi inc @ lw lw(-1)
creates equation EQ1 and calculates a LIML estimation of GDP on a constant, CPI, and INC, using a constant, LW, and LW(-1) as instruments.
e1.liml(kclass=2)
estimates the same equation, but this time via K-Class estimation, with K=2.
Cross-references
Estimate binary models with logistic errors.
Provide for backward compatibility. Equivalent to issuing the command, binary with the option “(d=l)”.
Estimation by linear or nonlinear least squares regression.
When the current workfile has a panel structure, ls also estimates cross-section weighed least squares, feasible GLS, and fixed and random effects models.
Syntax
eq_name.ls(options) y x1 [x2 x3 ...]
eq_name.ls(options) specification
For linear specifications, list the dependent variable first, followed by a list of the independent variables. Use a “C” if you wish to include a constant or intercept term; unlike some programs, EViews does not automatically include a constant in the regression. You may add AR, MA, SAR, and SMA error specifications, a D fractional differencing term, and PDL specifications for polynomial distributed lags. If you include lagged variables, EViews will adjust the sample automatically, if necessary.
Both dependent and independent variables may be created from existing series using standard EViews functions and transformations. EViews treats the equation as linear in each of the variables and assigns coefficients C(1), C(2), and so forth to each variable in the list.
Linear or nonlinear single equations may also be specified by explicit equation. You should specify the equation as a formula. The parameters to be estimated should be included explicitly: “C(1)”, “C(2)”, and so forth (assuming that you wish to use the default coefficient vector “C”). You may also declare an alternative coefficient vector using coef and use these coefficients in your expressions.
Options
Non-Panel LS Options
indicator | Include indicator saturation detection as part of estimation routine. |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
z | Turn off backcasting in ARMA models where “arma=cls”. |
optmethod = arg | Optimization method for nonlinear least squares and ARMA: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “kohn” (Kohn-Ansley for ARMA estimated by ML or GLS), or “legacy” (EViews legacy for nonlinear least squares and ARMA estimated by CLS). Gauss-Newton is the default method. |
optstep = arg | Step method for nonlinear least squares and ARMA: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method available for nonlinear least squares or ARMA estimated by CLS), “hac” (Newey-West HAC, available for nonlinear least squares or ARMA estimated by CLS).. |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 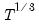 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
arma=arg | ARMA estimation method: “ml” (maximum likelihood); “gls” (generalized least squares), “cls” (conditional least squares). Not applicable to ARFIMA models which always estimate using maximum likelihood. |
armastart=arg | ARMA coefficient starting values: “auto” (automatic) “fixed” (legacy EViews fixed); “random” (random draw); “user” (user-specified). Applicable when “arma=ml” or “arma=gls”. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR or MA terms when “arma=cls” (see also
param ). |
s=number | Determine starting values for equations specified by list with AR or MA terms when “arma=cls”. Specify a number between zero and one representing the fraction of preliminary least squares estimates computed without AR or MA terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR and MA terms which are set to EViews determined default values. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. Available only for legacy estimation (“optmeth=legacy”). |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method available for nonlinear least squares or ARMA estimated by CLS), “hac” (Newey-West HAC, available for nonlinear least squares or ARMA estimated by CLS)., “hc” (extended heteroskedasticity consistent), “hcuser” (user-specified heteroskedasticity), “cr” (cluster robust). The extended “hc” methods are only available for linear specifications. |
hctype=arg (default “hc2”) | Extended heteroskedasticity consistent method: “hc0” (no d.f. adjustment), “hc1” (d.f. adjusted), “hc2”, “hc3”, “hc4”, “hc4m”, “hc5”, when “cov=hc”. |
userwt=arg | Name of series containing user-diagonal weights (if “cov=hcuser”) |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), “hc2”, “hc3”, “hc4”, “hc4m”, “hc5”, when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
k=arg (default = 0.7) | Parameter for “cov=hc, hctype=hc5” or “cov=cr, crtype=cr5”. |
k1=arg (default = 1.0) | Parameter for “cov=hc, hctype=hc4m” or “cov=cr, crtype=cr4m”. |
k2=arg (default = 1.5) | Parameter for “cov=hc, hctype=hc4m” or “cov=cr, crtype=cr4m”. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Note: not all options are available for all equation methods.
Non-Panel Indicator Saturation Options
For use if “indicator” option is specified.
noiis | Do not search for impulse terms. |
sis | Search for step-shift terms. |
trend | Search for trend terms. |
pval=number (default = 0.05) | Set the terminal condition p-value used to determine the stopping point of each search path |
nolm | Do not perform AR LM diagnostic test. |
arpval=number (default = 0.025) | Set p-value used in AR LM diagnostic test. |
arlags=int (default = 1) | Set number of lags used in AR LM diagnostic test. |
noarch | Do not perform ARCH LM diagnostic test. |
archpval=number (default = 0.025) | Set p-value used in ARCH LM diagnostic test. |
archlags=int (default = 1) | Set number of lags used in ARCH LM diagnostic test. |
nojb | Do not perform Jarque-Bera normality diagnostic test. |
jbpval=number (default = 0.025) | Set p-value used in Jarque-Bera normality diagnostic test. |
nopet | Do not perform Parsimonious Encompassing diagnostic test. |
petpval=number (default = 0.025) | Set p-value used in Parsimonious Encompassing diagnostic test. |
nogum | Do not include the general model as a candidate for model selection. |
noempty | Do not include the empty model as a candidate for model selection. |
ic =arg | Set the information criterion used in model selection: “AIC” (Akaike information criteria, default), “BIC” (Schwarz information criteria), “HQ” (Hannan-Quin criteria). |
blocks=int | Override the EViews’ determination of the number of blocks in which to split the estimation sample. |
Panel LS Options
cx=arg | Cross-section effects: (default) none, fixed effects (“cx=f”), random effects (“cx=r”). |
per=arg | Period effects: (default) none, fixed effects (“per=f”), random effects (“per=r”). |
wgt=arg | GLS weighting: (default) none, cross-section system weights (“wgt=cxsur”), period system weights (“wgt=persur”), cross-section diagonal weighs (“wgt=cxdiag”), period diagonal weights (“wgt=perdiag”). |
cov=arg | Coefficient covariance method: (default) ordinary, White cross-section system (period clustering) robust (“cov=cxwhite” or “cov=percluster”), White period system (cross-section clustering) robust (“cov=perwhite” or “cov=cxcluster”), White heteroskedasticity robust (“cov=stackedwhite”), White two-way cluster robust (cov=bothcluster”), Cross-section system robust/PCSE (“cov=cxsur”), Period system robust/PCSE (“cov=persur”), Cross-section heteroskedasticity robust/PCSE (“cov=cxdiag”), Period heteroskedasticity robust/PCSE (“cov=perdiag”). |
keepwgts | Keep full set of GLS weights used in estimation with object, if applicable (by default, only small memory weights are saved). |
rancalc=arg (default=“sa”) | Random component method: Swamy-Arora (“rancalc=sa”), Wansbeek-Kapteyn (“rancalc=wk”), Wallace-Hussain (“rancalc=wh”). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
iter=arg (default= “onec”) | Iteration control for GLS specifications: perform one weight iteration, then iterate coefficients to convergence (“iter=onec”), iterate weights and coefficients simultaneously to convergence (“iter=sim”), iterate weights and coefficients sequentially to convergence (“iter=seq”), perform one weight iteration, then one coefficient step (“iter=oneb”). Note that random effects models currently do not permit weight iteration to convergence. |
unbalsur | Compute SUR factorization in unbalanced data using the subset of available observations for a cluster. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR terms (see also
param ). |
s=number | Determine starting values for equations specified by list with AR terms. Specify a number between zero and one representing the fraction of preliminary least squares estimates computed without AR terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR terms which are instead set to EViews determined default values. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
equation eq1.ls m1 c uemp inf(0 to -4) @trend(1960:1)
estimates a linear regression of M1 on a constant, UEMP, INF (from current up to four lags), and a linear trend.
equation eq2.ls(z) d(tbill) c inf @seas(1) @seas(1)*inf ma(2)
regresses the first difference of TBILL on a constant, INF, a seasonal dummy, and an interaction of the dummy and INF, with an MA(2) error. The “z” option turns off backcasting.
coef(2) beta
param beta(1) .2 beta(2) .5 c(1) 0.1
equation eq3.ls(cov=white) q = beta(1)+beta(2)*(l^c(1) + k^(1-c(1)))
estimates the nonlinear regression starting from the specified initial values. The “cov=white” option reports heteroskedasticity consistent standard errors.
equation eq4.ls r = c(1)+c(2)*r(-1)+div(-1)^c(3)
sym betacov = eq4.@cov
declares and estimates a nonlinear equation and stores the coefficient covariance matrix in a symmetric matrix called BETACOV.
equation eq5.ls(cx=f, per=f) n w k ys c
estimates the equation EQ5 in the panel workfile using both cross-section and period fixed effects.
equation eq6.ls(cx=f, wgt=cxdiag) n w k ys c
estimates the equation EQ6 in a panel workfile with cross-section weights and fixed effects.
Cross-references
“Basic Regression Analysis” and
“Additional Regression Tools” discuss the various regression methods in greater depth.
“Special Expression Reference”describes special terms that may be used in
ls specifications.
See
“Panel Estimation”for a discussion of panel equation estimation.
Leverage plots.
Displays leverage plots to discover influential observations, or outliers.
Syntax
equation_name.lvageplot(options) variables @ name_suffix
where name_suffix is an optional naming suffix for storing the statistics into series in the workfile.
Options
raw | Do not use partial residuals. |
nofit | Do not include a line of fit on the graphs |
prompt | Force the dialog to appear from within a program. |
Examples
eq1.lvageplot x1 x2 @ lplot_
will display two graphs, one for the leverage plot of X1 and one for the leverage plot of X2, and will create two new series in the workfile, LPLOT_X1 and LPLOT_X2.
Cross-references
Create a series containing the estimated cointegrating relationship from an ARDL estimated equation.
This view is only available for non-panel equations estimated using the ARDL method.
Syntax
equation_name.makecoint [series_name]
Examples
wfopen http://www.stern.nyu.edu/~wgreene/Text/Edition7/TableF5-2.txt
equation eq02.ardl(deplags=3, reglags=3, fixed) log(realcons) log(realgdp) @ @expand(@quarter, @droplast)
show eq02.makecoint cointser
This example uses data from Greene (2008, page 685), containing quarterly US macroeconomic variables between 1950 and 2000. The first line of this example downloads the data set, the second line creates an equation object and estimates an ARDL model with the log of real consumption as the dependent variable. Three lags of the dependent variable, and three lags of the log of real GDP are used as dynamic regressors. Quarterly dummy variables are included as static regressors.
The final line creates a new series, COINTSER, containing the estimated cointegrating relationship.
Cross-references
See
“ARDL and Quantile ARDL” for further discussion.
Make a group containing individual series which hold the derivatives of the equation specification.
Syntax
equation_name.makederivs(options) [ser1 ser2 ...]
If desired, enclose the name of a new group object to hold the series in parentheses following the command name.
The argument specifying the names of the series is also optional. If not provided, EViews will name the series “DERIV##” where ## is a number such that “DERIV##” is the next available unused name. If the names are provided, the number of names must match the number of target series.
names must match the number of target series.
Options
n=arg | Name of group object to contain the series. |
Examples
eq1.makederivs(n=out)
creates a group named OUT containing series named DERIV01, DERIV02, and DERIV03.
eq1.makederivs(n=out) d1 d2 d3
creates the same group, but names the series D1, D2 and D3.
Cross-references
See
“State Space Models and the Kalman Filter” for details on state space estimation.
Export functional objects to matrices in the workfile.
For equations estimated using the functional coefficients method.
Syntax
eq_name.makefunobj(options) matrix_name
where matrix_name is the name of the output matrix.
Options
Basic Options
type=arg (default= “coef”) | Type of result to save: coefficients (“coef”), residuals (“res”), bias (“bias”), covariances involving a single coefficient (“cov”), correlations involving a single coefficient (“cor”), confidence intervals (“ci”). If saving covariances or correlations, you may identify the coefficient of interest using the “coefid=” option. |
wf | Make output of workfile length. Note: this option is only relevant if estimation was evaluated over the functional coefficient variable values. |
derivs | Include derivative coefficients as part of the output (for all but “type=resid”). Use derivative coefficients are part of the calculation (when type=resid”). |
nodups | Duplicate observations for in the set of functional coefficient evaluation points are removed. |
sort | Rows of the output matrix are sorted in increasing order of the functional coefficient evaluation points. |
Pilot Bandwidth Options
If a local pilot bandwidth has exists, all of the pilot bandwidth computation options below will be ignored unless the “nolocalbw” option is specified.
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
auxk=integer (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage degree. This number should always be an even positive integer. |
Covariance Options
coefid=integer (default = 1) | Coefficient ID for which covariances or correlations are produced when “type=cov” or “type=cov”. |
Confidence Interval Options
The following options are only available when “type=ci”.
seband | Produce standard error bands instead of confidence intervals. |
sewidth =integer (default = 1) | Number of standard errors to use as half-width of confidence band (when “seband” is specified). |
cilevel =arg (default = 0.95) | Confidence interval coverage as a number between 0 and 1 (not applicable when “seband” is specified). |
nobias | Ignore bias in confidence interval determination. |
Examples
eq.makefunobj funcoef
produces a matrix of functional coefficients (one column per coefficient) called “FUNCOEF” using the existing local pilot bandwidth if present, or estimating the pilot bandwidth, if not. If estimated, the local pilot bandwidth is updated with the result.
eq.makefunobj(type=cor, coefid=3) myfuncorr
produces a matrix of functional correlations with respect to the third functional coefficient. The matrix is stored in the workfile under the name “MYFUNCORR”.
eq.makefunobj(nolocalbw, type=ci, cilevel=0.9) myfunci
produces a matrix with the 90% functional confidence intervals. The lower and upper bounds of the intervals are paired in adjacent columns.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation. See
“Post-Estimation Views and Procs” for detail on the various functional calculations.
Generate conditional variance series.
Saves the estimated conditional variance (from an equation estimated using ARCH) as a named series.
Syntax
eq_name.makegarch series1_name [@ series2_name]
You should provide a name for the saved conditional standard deviation series following the makegarch keyword. If you do not provide a name, EViews will name the series using the next available name of the form “GARCH##” (if GARCH01 already exists, it will be named GARCH02, and so on).
For component GARCH equations, the permanent component portion of the conditional variance may be saved by adding “@” followed by a series name.
Options
prompt | Force the dialog to appear from within a program. |
Examples
equation eq1.arch sp c
eq1.makegarch cvar
plot cvar^.5
estimates a GARCH(1,1) model, saves the conditional variance as a series named
CVAR, and plots the conditional standard deviation. If you merely wish to view a plot of the conditional standard deviation without saving the series, use the
Equation::garch view.
The commands
equation eq1.arch(cgarch) sp c
eq1.makegarch cvar @ pvar
first estimates a Component GARCH model and then saves both the conditional variance and the permanent component portion of the conditional variance in the series CVAR and PVAR, respectively.
Cross-references
See
“ARCH and GARCH Estimation” for a discussion of GARCH models.
Make a group containing individual series which hold the gradients of the objective function.
Syntax
equation_name.makegrads(options) [ser1 ser2 ...]
The argument specifying the names of the series is also optional. If the argument is not provided, EViews will name the series “GRAD##” where ## is a number such that “GRAD##” is the next available unused name. If the names are provided, the number of names must match the number of target series.
Options
n=arg | Name of group object to contain the series. |
Examples
eq1.grads(n=out)
creates a group named OUT containing series named GRAD01, GRAD02, and GRAD03.
eq1.makegrads(n=out) g1 g2 g3
creates the same group, but names the series G1, G2 and G3.
Cross-references
Create vector of limit points from ordered models.
makelimits creates a vector of the estimated limit points from equations estimated by
Equation::ordered.
Syntax
eq_name.makelimits [vector_name]
Provide a name for the vector after the makelimits keyword. If you do not provide a name, EViews will name the vector with the next available name of the form LIMITS## (if LIMITS01 already exists, it will be named LIMITS02, and so on).
Examples
equation eq1.ordered edu c age race gender
eq1.makelimits cutoff
Estimates an ordered probit and saves the estimated limit points in a vector named CUTOFF.
Cross-references
See
“Ordered Dependent Variable Models” for a discussion of ordered models.
Make a model from an equation object.
Syntax
equation_name.makemodel(name) assign_statement
If you provide a name for the model in parentheses after the keyword, EViews will create the named model in the workfile. If you do not provide a name, EViews will open an untitled model window if the command is executed from the command line.
Examples
equation eq3.ls 1 4 m1 gdp tb3
eq3.makemodel(eqmod) @prefix s_
estimates an equation and makes a model named EQMOD from the estimated equation object. EQMOD includes an assignment statement “ASSIGN @PREFIX S_”. Use the command “show eqmod” or “eqmod.spec” to open the EQMOD window.
Cross-references
See
“Models” for a discussion of specifying and solving models in EViews. See also
solve.
Make regressor group.
Creates a group containing the dependent and independent variables from an equation specification.
Syntax
equation_name.makeregs grp_name
Follow the keyword makeregs with the name of the group.
Examples
equation eq1.ls y c x1 x2 x3 z
eq1.makeregs reggroup
creates a group REGGROUP containing the series Y X1 X2 X3 and Z.
Cross-references
Create residual series.
Creates and saves residuals in the workfile from an estimated equation object.
Syntax
equation_name.makeresids(options) [res1]
Follow the equation name with a period and the makeresids keyword, then provide a name to be given to the stored residual.
Options
o (default) | Ordinary residuals. |
s | Standardized residuals (available only after weighted estimation and GARCH, binary, ordered, censored, and count models). |
g (default for ordered models) | Generalized residuals (available only for binary, ordered, censored, and count models). |
prompt | Force the dialog to appear from within a program. |
Examples
equation eq1.ls y c m1 inf unemp
eq1.makeresids res_eq1
estimates a linear regression of Y on a constant, M1, INF, UNEMP, and saves the residuals as a series named RES_EQ1.
Cross-references
See
“Weighted Least Squares” for a discussion of standardized residuals after weighted least squares and
“Discrete and Limited Dependent Variable Models” for a discussion of standardized and generalized residuals in binary, ordered, censored, and count models.
Save the regime probabilities for switching regression equation into series in the workfile.
Syntax
equation_name.makergmprobs(options) series_names
where equation_name is the name of an equation estimated using switching regression. The series to be saved should be listed following the command name and options, with one name per regime for one up to the number of estimated regimes.
Options
type=arg (default=“pred”) | Type of regime probability to compute: one-step ahead predicted (“pred”), filtered (“filt”), smoothed (“smooth”). |
n=arg | (optional) Name of group to contain the saved regime probabilities. |
prompt | Force the dialog to appear from within a program. |
Examples
equation eq1.switchreg(type=markov) y c @nv ar(1) ar(2) ar(3)
eq1.makergmprobs r1 r2
saves the one-step ahead regime probabilities for the Markov switching regression estimated in EQ1 in series R1 and R2 in the workfile
eq1.makergmprobs(type=filt) f1
saves the filtered probabilities for regime 1 in F1.
eq1.makergmprobs(type=smooth, n=smoothed) s1 s2
saved the smoothed probabilities for both regimes in the series S1 and S2, and creates the group SMOOTHED containing S1 and S2.
Cross-references
Save a series containing the smooth transition weights for each observation in the estimation sample in a smooth threshold regression.
Syntax
eq_name.makestrwgts(options) name
The command makes weight series in the workfile using name or the next available name using name as a basename.
Options
prompt | Force the dialog to appear from within a program. |
Examples
eq1.makestrwgts w
saves the weights in the series W.
Cross-references
Save the regime transition probabilities and expected durations for a switching regression equation into the workfile.
Syntax
equation_name.maketransprobs(options) [base_name]
equation_name.maketransprobs(out=mat, options) [matrix_name]
where equation_name is the name of an equation estimated using switching regression.
• In the first form of the command,
base_name will be used to generate series names for the series that will hold the transition probabilities or durations. The series names for regime transition probabilities will be of the form
base_name##, where ## are the indices representing elements of the transition matrix
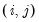
. The series names for expected durations will be of the form
base_name# where # corresponds to the regime index. Thus, in a two-regime model, the base name “TEMP” corresponds to the transition probability series TEMP11, TEMP12, TEMP21, TEMP22, and the expected duration series TEMP1, TEMP2.
If base_name is not provided, EViews will use the default of “TPROB”
• When the option “output=mat” is provided, the matrix_name is the name of the output matrix that will hold the transition probabilities or durations.
If matrix_name are not provided, EViews will default to “TPROB” or the next available name of the form “TPROB##”.
EViews will evaluate the transition probabilities or durations at the date specified by the “obs=” option. If no observation is specified, EViews will use the first date of the estimation sample to evaluate the transition probabilities. Note that if the transition probabilities are time-invariant, setting the observation will have no effect on the contents of the saved results.
Options
type=arg (default=“trans”) | Transition probability results to save: transition probabilities (“trans”), expected durations (“expect”). |
out=arg (default=“series”) | Output format: series (“series”) or matrix (“mat”). If saved as a matrix, only a single transition matrix will be saved using the date specified by “obs=”. |
obs=arg | Date/observation used to evaluate the transition probabilities if saving results as a matrix (“out=mat”). If no observation is specified, EViews will use the first date of the estimation sample to evaluate the transition probabilities. Note that if the transition probabilities are time-invariant, setting the observation will have no effect on the content of the saved results. |
n=arg | (optional) Name of group to contain the saved transition probabilities. |
prompt | Force the dialog to appear from within a program. |
Examples
equation eq1.switchreg(type=markov) y c @nv ar(1) ar(2) ar(3)
eq1.maketransprobs(n=transgrp) trans
saves the transition probabilities in the workfile in the series TRANS11, TRANS12, TRANS21, TRANS22 and creates the group TRANSGRP containing the series.
The command
eq1.maketransprobs(type=expect) AA
saves the expected durations in the series AA1 and AA2.
eq1.maketransprobs(out=mat) BB
saves the transition probabilities in the matrix BB.
Cross-references
Descriptive statistics by category of dependent variable.
Computes and displays descriptive statistics of the explanatory variables (regressors) of an equation categorized by values of the dependent variable for binary and censored/truncated models
Syntax
eq_name.means(options)
Options
p | Print the descriptive statistics table. |
Examples
equation eq1.binary(d=l) work c edu faminc
eq1.means
estimates a logit and displays the descriptive statistics of the regressors C, EDU, FAMINC for WORK=0 and WORK=1.
Cross-references
See
“Discrete and Limited Dependent Variable Models” for a discussion of binary and censored/truncated dependent variable models.
Estimates an equation using Mixed Data Sampling (MIDAS) regression.
MIDAS regression is an estimation technique which allows for data sampled at different frequencies to be used in the same regression.
Syntax
eq_name.midas(options) y x1 [x2 x3 ...] @ z1page\z1 [z2page\z2 ...]
where y, x1, etc., are the dependent and explanatory variables in the current page frequency, and z1page\z1 and z2page\z2 are the high frequency variable page\series specification.
You may not include ARMA terms in a MIDAS regression.
Options
General options
midwgt=arg | MIDAS weight method: step function(“step”), normalized exponential Almon (“expalmon”), normalized beta function (“beta”), U-MIDAS (“umidas”), Auto-search/GETS (“autogets”) or the default Almon/PDL weighting (“almon”). |
lag=arg | Method for specifying the number of lags of the high frequency regressor to use: lag selection (“auto”), fixed (“fixed”). The default is “lag=fixed”. |
maxlag=arg | Maximum number of lags of the high frequency regressor to use when using lag selection. For use when “lag=auto”. The default value is 4. |
fixedlag=arg | Fixed number of lags of the high frequency regressor to use. For use when “lags=fixed”. The default value is 4. |
steps=integer | Stepsize (number of high frequency periods to group). For use when “midwgt=step”. |
polynomial=integer | Polynomial degree. For use when Almon/PDL weighting is employed. |
beta=arg | Beta function restriction: none (“none”), trend coefficient equals 1 (“trend”), endpoints coefficient equals 0 (“endpoint”), both trend and endpoints restriction (“both”). For use when “midwgt=beta”. The default is “beta=none”. |
optmethod = arg | Optimization method for nonlinear estimation: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “hybrid” (initial BHHH followed by BFGS). Hybrid is the default method. |
optstep = arg | Step method for nonlinear estimation: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method for nonlinear models: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich). |
covinfo = arg | Information matrix method for nonlinear models: “opg” (OPG); “hessian” (observed Hessian). |
freq = key | Set the frequency conversion method. Key can be “first” (the higher frequency data are used from the first observation in the lower frequency period), “last” (default, the higher frequency data are used from the last observation in the lower frequency), or “match” (a specific date matching series from each page is used). |
freqsrc = arg | Set the source date matching series. Only applies if freq=match is used. |
freqdest = arg | Set the destination date matching series. Only applies if freq=match is used. |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values in nonlinear estimation (see also
param). |
s=number | Determine starting values for nonlinear estimation.. Specify a number between zero and one representing the fraction of preliminary EViews chosen values. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Auto-search/GETS options
pval=number (default = 0.05) | Set the terminal condition p-value used to determine the stopping point of each search path |
nolm | Do not perform AR LM diagnostic test. |
arpval=number (default = 0.025) | Set p-value used in AR LM diagnostic test. |
arlags=int (default = 1) | Set number of lags used in AR LM diagnostic test. |
noarch | Do not perform ARCH LM diagnostic test. |
archpval=number (default = 0.025) | Set p-value used in ARCH LM diagnostic test. |
archlags=int (default = 1) | Set number of lags used in ARCH LM diagnostic test. |
nojb | Do not perform Jarque-Bera normality diagnostic test. |
jbpval=number (default = 0.025) | Set p-value used in Jarque-Bera normality diagnostic test. |
nopet | Do not perform Parsimonious Encompassing diagnostic test. |
petpval=number (default = 0.025) | Set p-value used in Parsimonious Encompassing diagnostic test. |
nogum | Do not include the general model as a candidate for model selection. |
noempty | Do not include the empty model as a candidate for model selection. |
ic =arg | Set the information criterion used in model selection: “AIC” (Akaike information criteria, default), “BIC” (Schwarz information criteria), “HQ” (Hannan-Quin criteria). |
blocks=int | Override the EViews’ determination of the number of blocks in which to split the estimation sample. |
Examples
equation eq1.midas(fixedlag=9, midwgt=beta, beta=endpoint) realgdp c realgdp(-1) @ monthlypage\emp(-5)
estimates a MIDAS beta weight specification using the low frequency dependent variable REALGDP and regressors C and REALGDP(-1), and 9 beta weighted lags of EMP(-5) from the “monthlypage” workfile page. The beta weight function places zero restrictions on the endpoint coefficient.
equation eq2.midas(maxlag=12, lag=auto) realgdp c realgdp(-1) @ monthlypage\emp(-5)
estimates the same equation using PDL/Almon weights. The number of lags is chosen automatically with a maximum of 12 lags.
Cross-references
“Midas Regression” discusses the specification and estimation of MIDAS regression models in EViews.
Display a graph of the selection criteria for the top 20 models as determined by model selection during estimation.
This view is currently only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
The graph elements will be ordered by the value of the selection criterion.
Syntax
equation_name.modselgraph
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.modselgraph
shows a plot of the top 20 model selection objective values.
Cross-references
For further discussion, see
“Elastic Net and Lasso”The data underlying this graph are available via the data member @modselresults.
Display the selection objective and fit measures associated with the estimation and model selection procedures.
This view is only available for equations estimated with elastic net, ridge regression, Lasso, and variable selection using Lasso.
Displays a table showing the model selection objective, number of non-zero coefficients, and the fit statistics (sum-of-squared residuals, mean-square error, R-squared, and adjusted R-squared) associated with the estimated model.
The row displaying the model selected optimal lambda will be highlighted.
Syntax
eq_name.modseltable(options)
Options
Examples
Consider the estimated elastic net equation
equation my_eq.enet(xtrans=none, lambdaratio=.0001, cvseed=513255899) lpsa c lcavol_s lweight_s age_s lbph_s svi_s lcp_s gleason_s pgg45_s
Then the command
my_eq.modseltable
displays the table showing the model selection objective and various fit statistics associated with each lambda in the path.
Cross-references
For further discussion, see
“Elastic Net and Lasso”.
The data underlying this table are available via the data member @modselresults.
Multiple breakpoint testing.
The multibreak view of an equation displays the results of multiple breakpoint testing using sequential and global optimization methods.
This view is only available for (non-panel) equations specified by list without ARMA terms and estimated by ordinary least squares.
Syntax
equation_name.multibreak(options) [list_of_breaking_regressors]
where equation_name is the name on an equation specified by list and estimated using least squares. The multibreak may be followed by options, and an optional list of breaking regressor names. If the latter list is omitted, the coefficients for all of the regressors in the original equation will be allowed to vary across regimes.
Options
method=arg (default=“seqplus1”) | Breakpoint testing method: “seqplus1” (sequential tests of single 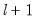 versus 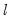 breaks), “seqall” (sequential test of all possible 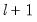 versus 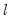 breaks), “glob” (tests of global 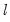 vs. no breaks), “globplus1” (tests of 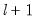 versus 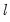 globally determined breaks), “globinfo” (information criteria evaluation). |
trim=arg (default=5) | Trimming percentage for determining minimum segment size (5, 10, 15, 20, 25). |
maxbreaks=integer (default=5) | Maximum number of breakpoints to allow (not applicable if “method=seqall”). |
maxlevels=integer (default=5) | Maximum number of break levels to consider in sequential testing (applicable when “method=sequall”). |
size=arg (default=5) | Test sizes for use in sequential determination and final test evaluation (10, 5, 2.5, 1) corresponding to 0.10, 0.05, 0.025, 0.01, respectively |
heterr | Assume regimes specific error distributions in variance computation. |
commondata | Assume a common distribution for the data across segments (only applicable if original equation is estimated with a robust covariance method, “heterr” is not specified). |
prompt | Force the dialog to appear from within a program. |
p | Print the view. |
Examples
equation eq01.ls m1 c tb3 gdp
eq01.multibreak(maxbreaks=3)
eq01.multibreak(method=glob, size=10, trim=15) tb3
The first test line tests for up to 3 structural breaks in all of the coefficients using sequential tests of single
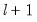
versus
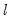
breaks. The second line tests uses the global
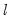
breaks versus none tests with trimming value 0.15, and a size of 0.10 to test for differences in the coefficient on TB3 across regimes.
The multiple breakpoint tests will use the covariance matrix settings from the original equation when constructing tests. The command
equation eq01.ls(cov=hac, covkern=quadspec, covlag=1, covbw=andrews) rates c
eq01.multibreak(heterr)
eq01.multibreak(method=glob, heterr)
eq01.multibreak(method=globinfo)
estimate an equation using HAC covariances. The second line tests for up to 5 structural breaks using sequential tests of single
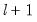
versus
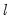
breaks. The third line uses the global
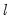
breaks versus none tests. Both of these tests allow for error distributions to vary across the different segments. The final line evaluates the breakpoints using information criteria associated with the global optimizers.
Cross-references
See
“Multiple Breakpoint Tests” for discussion. See also
“Least Squares with Breakpoints” for tools which estimate equations with structural breaks.
See
Equation::breakls for estimation of regression equations with breaks.
Display a news-impact graph of equations estimated using GARCH.
Syntax
eq_name.newsimpact(options)
Options
Examples
equation eq1.arch(2, 1) y c
estimates a GARCH(2, 1) model.
eq1.newsimpact
displays the news-impact graph of the estimated GARCH model.
Cross-references
Perform the Nyblom test of parameter stability or structural change in equations estimated using GARCH.
Syntax
eq_name.nyblom(options)
Options
Examples
equation eq1.arch(2, 1) y c
estimates a GARCH(2, 1) model.
eq1.nyblom
displays the results of a Nyblom stability test.
Cross-references
Push updates to OLE linked objects in open applications.
Syntax
equation_name.olepush
Cross-references
See
“Object Linking and Embedding (OLE)” for a discussion of using OLE with EViews.
Estimation of ordered dependent variable models.
Syntax
equation name.ordered(options) y x1 [x2 x3 ...]
equation name.ordered(options) specification
The ordered command estimates the model and saves the results as an equation object with the given name.
Options
d=arg (default=“n”) | Specify likelihood: normal likelihood function, ordered probit (“n”), logistic likelihood function, ordered logit (“l”), Type I extreme value likelihood function, ordered Gompit (“x”). |
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). Newton-Raphson is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method)., “glm” (GLM method), “cr” (cluster robust). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
df | Degree-of-freedom correct the coefficient covariance estimate.(For non-cluster robust methods estimated using non-legacy estimation). |
h | Huber-White quasi-maximum likelihood (QML) standard errors and covariances. (Legacy option Applicable when “optmethod=legacy”). |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of preliminary EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
If you choose to employ user specified starting values, the parameters corresponding to the limit points must be in ascending order.
Examples
ordered(d=l,cov=huber) y c wage edu kids
estimates an ordered logit model of Y on a constant, WAGE, EDU, and KIDS with QML standard errors. This command uses the default quadratic hill climbing algorithm.
param c(1) .1 c(2) .2 c(3) .3 c(4) .4 c(5).5
equation eq1.binary(s) y c x z
coef betahat = eq1.@coefs
eq1.makelimit gamma
estimates an ordered probit model of Y on a constant, X, and Z from the specified starting values. The estimated coefficients are then stored in the coefficient vector BETAHAT, and the estimated limit points are stored in the vector GAMMA.
Cross-references
See
Equation::binary for the estimation of binary dependent variable models. See also
Equation::makelimits.
Performs the Instrument Orthogonality Test
The Orthogtest view of an equation carries out the Instrument Orthogonality / C-test Test for equations estimated via TSLS or GMM.
Syntax
eq_name.orthogtest(options) instruments
Options
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Instruments
A list of instruments to be tested for orthogonality. Note the instruments must have been included in the original equation.
Examples
equation eq1.gmm y c x1 x2 @ z1 z2 z3 z4
e1.orthogtest z1 z4
estimates an equation, called EQ1, and estimates it via GMM with four instruments Z1, Z2, Z3, Z4, and then performs the Orthogonality Test where Z1 and Z4 are tested for orthogonality.
Cross-references
Display the outliers summary view for an equation estimated via least squares with automatic outlier indicator saturation.
Syntax
equation_name.outliers
Examples
equation eq1.ls(indicator) y c x
show eq1.outliers
Estimates a least squares regression with Y as the dependent variable, and X as a regressor, with an intercept, and with automatic detection of outliers via indicator saturation, and then displays the summary of the outlier detection routine.
Display estimation output.
The
output command changes the default object view to display the equation output (equivalent to using
Equation::results).
Syntax
eq_name.output(options)
Options
p | Print estimation output for estimation object. |
Examples
eq1.output
displays the estimation output for equation EQ1.
Cross-references
Display the auto-gets paths view.
Syntax
equation_name.paths
Examples
eq1.varsel(method=gets) y c @ x1 x2 x3 x4 x5 x6 x7 x8
show eq1.paths
performs a variable selection routine using the auto-gets method, and then displays the paths view of that procedure.
Cross-references
See also
“Regression Variable Selection”for extensive discussion.Displays a spool object with the results of the Hausman test for similarity against mean-group and dynamic fixed effects estimators in PMG estimation.
Syntax
equation_name.pmghausmantest(options)
Options
Examples
equation eq.ardl log(cons) log(inf)
eq.pmghausmantest
Displays a spool object with the results of the Hausman test for similarity against mean-group and dynamic fixed effects estimators in PMG estimation.
Cross-references
Prediction table for binary and ordered dependent variable models.
The prediction table displays the actual and estimated frequencies of each distinct value of the discrete dependent variable.
Syntax
eq_name.predict(n, options)
For binary models, you may optionally specify how large the estimated probability must be to be considered a success (
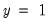
). Specify the cutoff level as the first option in parentheses after the keyword
predict.
Options
n (default=.5) | Cutoff probability for success prediction in binary models (between 0 and 1). |
prompt | Force the dialog to appear from within a program. |
p | Print the prediction table. |
Examples
equation eq1.binary(d=l) work c edu age race
eq1.predict(0.7)
Estimates a logit and displays the expectation-prediction table using a cutoff probability of 0.7.
Cross-references
Estimation of binary dependent variable models with normal errors.
Equivalent to “binary(d=n)”.
Displays a spool object producing a quantile process of the cointegrating relation.
Syntax
eq_name.qrcrprocess(options) [arg]
where arg is an optional list containing the quantile values (specified using numbers, scalar objects, or vectors) for which you wish to compute estimates.
• If arg is not specified, EViews will display results for the original equation along with coefficients for equations estimated at a set of equally spaced number of quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the deciles.
• If arg is specified, EViews will display results for the original equation along with coefficients for equations estimated at the specified quantiles.
Options
n=arg (default=10) | Number of quantiles for process estimates. |
quantout=name | Save vector containing test quantile values. |
coefout=name | Save matrix containing coefficient estimates of the cointegrating relation form. Each column of the matrix corresponds to a different quantile matching the corresponding quantile in "quantout=". To match the covariance matrix given in "covout=" you should take the @vec of the coefficient matrix. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
ardl_eq.qrcrprocess
This generates a quantile process of the cointegrating relation for each quantile running from 0.1 to 0.9.
ardl_eq.qrcrprocess(n=4, quantout=_vec_q, coefout=_mat_cointrel)
ardl_eq.qrcrprocess(quantout=_vec_q, coefout=_mat_cointrel) 0.25 0.5 0.75
Both commands generate the cointegrating relation process for each of the quantile values 0.25, 0.5, and 0.75. Additionally, this produces 2 workfile objects, "_vec_q" (a vector), "_mat_cointrel¨(a matrix), and "_mat_cointrel" (a matrix), corresponding to the three quantile values and the cointegrating relation process.
Cross-references
Displays a spool object producing a quantile process for each of the conditional error correction and error correction coefficients.
Syntax
eq_name.qrecprocess(options) [arg]
where arg is a optional list containing the quantile values (specified using numbers, scalar objects, or vectors) for which you wish to compute estimates.
• If arg is not specified, EViews will display results for the original equation along with coefficients for equations estimated at a set of equally spaced number of quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the deciles.
• If arg is specified, EViews will display results for the original equation along with coefficients for equations estimated at the specified quantiles.
Options
n=arg (default=10) | Number of quantiles for process estimates. |
quantout=name | Save vector containing test quantile values. |
coefout=name | Saves two matrices differentiated by suffixes "_cec" and "_ec", corresponding to the coefficients associated with the CEC and EC forms, respectively. Each column of the matrix corresponds to a different quantile matching the corresponding quantile in "quantout=". To match the covariance matrix given in "covout=" you should take the @vec of the coefficient matrix. |
covout=name | Saves two symmetric matrices, differentiated by suffixes "_cec" and "_ec", corresponding to the covariance matrices of coefficients associated with the CEC and EC forms, respectively. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
ardl_eq.qrecprocess
This generates a quantile process of the CEC and EC coefficients for each quantile running from 0.1 to 0.9.
ardl_eq.qrecprocess(n=4, quantout=_vec_q, coefout=_mat_coefs, covout=_mat_covs)
ardl_eq.qrecprocess(quantout=_vec_q, coefout=_mat_coefs, covout=_mat_covs) 0.25 0.5 0.75
Both commands generate the process for CEC and EC coefficients for each of the quantile values 0.25, 0.5, and 0.75. Additionally, this produces 5 workfile objects, "_vec_q" (a vector), "_mat_coefs_cec" (a matrix), "_mat_coefs_ec" (a matrix), "_mat_covs_cec" (a matrix), and "_mat_covs_ec" (a matrix), corresponding to the three quantile values, coefficient process for each of the CEC and EC forms, covariances process for each of the CEC and EC forms, respectively.
Cross-references
Estimate a quantile regression specification.
Syntax
eq_name.qreg(options) y x1 [x2 x3 ...]
eq_name.qreg(options) linear_specification
Options
quant=number (default = 0.5) | Quantile to be fit (where number is a value between 0 and 1). |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=arg (default=“sandwich”) | Method for computing coefficient covariance matrix: “iid” (ordinary estimates), “sandwich” (Huber sandwich estimates), “boot” (bootstrap estimates). When “cov=iid” or “cov=sandwich”, EViews will use the sparsity nuisance parameter calculation specified in “spmethod=” when estimating the coefficient covariance matrix. |
bwmethod=arg (default = “hs”) | Method for automatically selecting bandwidth value for use in estimation of sparsity and coefficient covariance matrix: “hs” (Hall-Sheather), “bf” (Bofinger), “c” (Chamberlain). |
bw =number | Use user-specified bandwidth value in place of automatic method specified in “bwmethod=”. |
bwsize=number (default = 0.05) | Size parameter for use in computation of bandwidth (used when “bw=hs” and “bw=bf”). |
spmethod=arg (default=“kernel”) | Sparsity estimation method: “resid” (Siddiqui using residuals), “fitted” (Siddiqui using fitted quantiles at mean values of regressors), “kernel” (Kernel density using residuals) Note: “spmethod=resid” is not available when “cov=sandwich”. |
btmethod=arg (default= “pair”) | Bootstrap method: “resid” (residual bootstrap), “pair” (xy-pair bootstrap), “mcmb” (MCMB bootstrap), “mcmba” (MCMB-A bootstrap). |
btreps=integer (default=100) | Number of bootstrap repetitions |
btseed=positive integer | Seed the bootstrap random number generator. If not specified, EViews will seed the bootstrap random number generator with a single integer draw from the default global random number generator. |
btrnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator for the bootstrap: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
btobs=integer | Number of observations for bootstrap subsampling (when “bsmethod=pair”). Should be significantly greater than the number of regressors and less than or equal to the number of observations used in estimation. EViews will automatically restrict values to the range from the number of regressors and the number of estimation observations. If omitted, the bootstrap will use the number of observations used in estimation. |
btout=name | (optional) Matrix to hold results of bootstrap simulations. |
k=arg (default=“e”) | Kernel function for sparsity and coefficient covariance matrix estimation (when “spmethod=kernel”): “e” (Epanechnikov), “r” (Triangular), “u” (Uniform), “n” (Normal–Gaussian), “b” (Biweight–Quartic), “t” (Triweight), “c” (Cosinus). |
m=integer | Maximum number of iterations. |
s | Use the current coefficient values in estimator coefficient vector as starting values (see also
param). |
s=number (default =0) | Determine starting values for equations. Specify a number between 0 and 1 representing the fraction of preliminary least squares coefficient estimates. Note that out of range values are set to the default. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
equation eq1.qreg y c x
estimates the default least absolute deviations (median) regression for the dependent variable Y on a constant and X. The estimates use the Huber Sandwich method for computing the covariance matrix, with individual sparsity estimates obtained using kernel methods. The bandwidth uses the Hall and Sheather formula.
equation eq1.qreg(quant=0.6, cov=boot, btmethod=mcmba) y c x
estimates the quantile regression for the 0.6 quantile using MCMB-A bootstrapping to obtain estimates of the coefficient covariance matrix.
Cross-references
See
“Quantile Regression” for a discussion of the quantile regression.
Display quantile process coefficient estimates (multiple quantile regression estimates).
Syntax
eq_name.qrprocess(options) [arg] [@coefs coeflist]
where arg is a optional list containing the quantile values (specified using numbers, scalar objects, or vectors) for which you wish to compute estimates, and optionally the @coefs keyword followed by a coeflist of the subset of coefficients to display.
• If arg is not specified, EViews will display results for the original equation along with coefficients for equations estimated at a set of equally spaced number of quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the deciles.
• If arg is specified, EViews will display results for the original equation along with coefficients for equations estimated at the specified quantiles.
• If a coeflist is not provided, results for all coefficients will be displayed. For models that contain an intercept, the coeflist may consist of the @incptonly keyword, indicating that only results for the intercept will be displayed.
You may specify a maximum of 1000 total coefficients (number of display coefficients times the number of quantiles) and a maximum of 500 quantiles.
All estimation will be performed using the settings from the original equation.
Options
n=arg (default=10) | Number of quantiles for process estimates. |
graph | Display process estimate results as graph. |
size=arg (default=0.95) | Confidence interval size for graph display |
quantout=name | Save vector containing test quantile values. |
coefout=name | Save matrix containing test coefficient estimates. Each column of the matrix corresponds to a different quantile matching the corresponding quantile in “quantout=”. To match the covariance matrix given in “covout=” you should take the @vec of the coefficient matrix. |
covout=name | Save symmetric matrix containing covariance matrix for the vector set of coefficient estimates. |
prompt | Force the dialog to appear from within a program. |
p | Print output. |
Examples
equation eq1.qreg log(y) c log(x)
eq1.qrprocess
estimates a quantile (median) regression of LOG(Y) on a constant and LOG(X), and displays results for all nine quantiles in a table
Similarly,
equation eq1.qreg(quant=.4) log(y) c log(x)
eq1.qrprocess(coefcout=cout)
displays the coefficient estimated at the deciles (and at 0.4), and saves the coefficient matrix to COUT.
eq1.qrprocess(coefout=cout, n=4, graph)
eq1.qrprocess(coefout=cout, graph) .25 .5 .75
both estimate coefficients for the three quartiles and display the results in a graph, as does the equivalent:
vector v1(3)
v1.fill .25 .5 .75
eq1.qrprocess(graph) v1
Cross-references
See
“Process Coefficients” for a discussion of the quantile process.
Perform Wald test of equality of slope coefficients across multiple quantile regression estimates. The equality test restrictions are of the form:
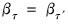
for the slope coefficients
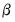
.
Syntax
eq_name.qrslope(options) [arg] [@coefs coeflist]
where arg is a optional list containing the quantile values (specified using numbers, scalar objects, or vectors) for which you wish to compute estimates, and optionally the @coefs keyword followed by a coeflist of the subset of coefficients to display.
• If arg is not specified, EViews will perform tests for the existing equation and coefficients for equations estimated at a set of equally spaced quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the quartiles (.25, .75).
• If arg is specified, EViews will perform results for the original equation along with tests including coefficients for equations estimated at the specified quantiles.
• If a coeflist is not provided, all of the slope coefficients will be employed in the test.
You may specify a maximum of 1000 total coefficients (number of coefficients in the equation specification times the number of quantiles) and a maximum of 500 quantiles in the test.
All estimation will be performed using the settings from the original equation.
Options
n=arg (default=4) | Number of quantiles for process estimates. |
quantout=name | Save vector containing test quantile values. |
coefout=name | Save matrix containing test coefficient estimates. Each column of the matrix corresponds to a different quantile matching the corresponding quantile in “quantout=”. To match the covariance matrix given in “covout=” you should take the @vec of the coefficient matrix. |
covout=name | Save symmetric matrix containing covariance matrix for the vector set of coefficient estimates. |
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
equation eq1.qreg log(y) c log(x)
eq1.qrslope
estimates a quantile (median) regression of LOG(Y) on a constant and LOG(X), and tests for the equality of the coefficients of LOG(X) for all three of the quartiles.
Similarly,
equation eq1.qreg(quant=.4) log(y) c log(x)
eq1.qrslope(coefcout=cout)
tests for equality of the LOG(X) coefficient estimated at {.25, .4, .5, .75}, and saves the coefficient matrix to COUT. Both
eq1.qrslope(coefout=count, n=10)
eq1.qrslope(coefout=cout) .1 .2 .3 .4 .5 .6 .7 .8 .9
perform the Wald test for equality of the slope coefficient across all of the deciles, as does the equivalent
vector v1(9)
v1.fill .1,.2,.3,.4,.5,.6,.7,.8,.9
eq1.qrslope v1
Cross-references
See
“Slope Equality Test” for a discussion of the slope equality test.
Perform Wald test of coefficients using symmetric quantiles. The symmetric quantile test restrictions are of the form:
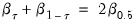
.
Syntax
eq_name.qrsymm(options) [arg] [@coefs coeflist]
where arg is a optional list containing the quantile values (specified using numbers, scalar objects, or vectors) for which you wish to compute estimates, and optionally the @coefs keyword followed by a coeflist of the subset of coefficients to display.
• If arg is not specified, EViews will perform one of two tests, depending on the original equation specification:
If the original specification is a median regression (
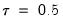
), EViews will test using estimates obtained at the specified outer quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the outer quartiles {0.25, 0.75}.
For specifications estimated with
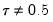
, EViews will include the original quantile in the set of quantiles to test. You may specify “n=e” to perform a test using only estimates obtained at the symmetric pair {
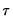
,
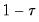
}.
• If arg is specified, EViews will perform the test using only the specified quantiles and their complements. The original equation quantile will not be tested unless it is entered explicitly.
• If a coeflist is not provided, results for all coefficients will be displayed. For models that contain an intercept, the coeflist may consist of the @incptonly keyword, indicating that only results for the intercept will be displayed.
You may specify a maximum of 1000 total coefficients (number of coefficients in the equation specification times the number of quantiles) and a maximum of 500 quantiles in the test.
All estimation will be performed using the settings from the original equation. Note that the original equation must include an intercept for you to perform this test
Options
n=arg (default=4) | Number of quantiles for testing. |
quantout=name | Save vector containing test quantile values. |
coefout=name | Save matrix containing test coefficient estimates. Each column of the matrix corresponds to a different quantile matching the corresponding quantile in “quantout=”. To match the covariance matrix given in “covout=” you should take the @vec of the coefficient matrix. |
covout=name | Save symmetric matrix containing covariance matrix for the vector set of coefficient estimates. |
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
equation eq1.qreg log(y) c log(x)
eq1.qrsymm
estimates a quantile (median) regression of LOG(Y) on a constant and LOG(X), and performs a symmetry test using the outer quartiles.
We may restrict the hypothesis to just consider the intercept,
eq1.qrsymm @coefs @incptonly
and we may specify alternative quantiles to test
eq1.qrsymm(quantout=qo) .2 .4 .7
Note that the latter command will test using the symmetric quantiles {0.2, 0.3, 0.4, 0.6, 0.7, 0.8}, and at the median. Note that the median is automatically estimated, even though it is not specified explicitly, since it is always required for testing.
Alternatively, the commands
equation eq1.qreg(quant=.4) log(y) c log(x)
eq1.qrsymm(n=0)
will perform the test using the symmetric quantiles {0.4, 0.6} and the median.
To performs the test using all of the deciles, you may enter
vector(4) v1
v1.fill .1,.2,.3,.4
eq1.qrsymm v1
Cross-references
See
“Symmetric Quantiles Test” for a discussion of the symmetric quantiles test.
Test for correlation between random effects and regressors using Hausman test.
Tests the hypothesis that the random effects (components) are correlated with the right-hand side variables in a panel or pool equation setting. Uses Hausman test methodology to compare the results from the estimated random effects specification and a corresponding fixed effects specification. If the estimated specification involves two-way random effects, three separate tests will be performed; one for each set of effects, and one for the joint effects.
Only valid for panel or pool regression equations estimated with random effects. Note that the test results may be suspect in cases where robust standard errors are employed.
Syntax
eq_name.ranhaus(options)
Options
p | Print output from the test. |
Examples
equation eq1.ls(cx=r) sales c adver lsales
eq1.ranhaus
estimates a specification with cross-section random effects and tests whether the random effects are correlated with the right-hand side variables ADVER and LSALES using the Hausman test methodology.
Cross-references
Tests for the presence of cross-sectional or time random components in a panel equation. estimated using pooled least squares.
Computes the conventional LM (Breusch-Pagan, 1980, uniformly most powerful LM (Honda, 1985), standardized Honda (Moulton and Randolph, 1989; Baltagi, Chang, and Li, 1998), locally mean most powerful (LMMP) (King and Wu, 1997), Standardized King-Wu, and Gourieroux, Holly, and Monfort (1982) test statistics.
Note that the equation must be estimated with pooled least squares for this test to be applied.
Syntax
equation_name.rcomptest
Options
G
Examples
equation eq1.ls @log(gsp) c @log(p_cap) @log(pc) @log(emp) unemp
eq1.rcomptest
will estimate a panel model using pooled least squares and will compute and display the panel random effects test results.
Cross-references
Display text of specification for equation objects.
Syntax
equation_name.representation(options)
Options
p | Print the representation text. |
Examples
eq1.representations
displays the specifications of the equation object EQ1.
Cross-references
Compute Ramsey’s regression specification error test.
Syntax
eq_name.reset(n, options)
You must provide the number of powers of fitted terms n to include in the test regression.
Options
prompt | Force the dialog to appear from within a program. |
p | Print the test result. |
Examples
equation eq1.ls lwage c edu race gender
eq1.reset(2)
carries out the RESET test by including the square and the cube of the fitted values in the test equation.
Cross-references
See
“Ramsey's RESET Test” for a discussion of the RESET test.
Display residuals.
The resids command allows you to display the actual, fitted values and residuals in either tabular or graphical form.
Syntax
equation_name.resids(options)
Options
g (default) | Display graph of actual/fittted/residuals (with one standard error bands) |
n | Display graph of residuals only (with one standard error bands) |
t | Display table of actual/fitted/residuals. |
s | Display graph of standardized residuals. |
p | Print the table/graph. |
Examples
equation eq1.ls m1 c inc tb3 ar(1)
eq1.resids
regresses M1 on a constant, INC, and TB3, correcting for first order serial correlation, and displays a table of actual, fitted, and residual series.
eq1.resids(g)
displays a graph of the actual, fitted, and residual series.
Cross-references
Detect outliers in the residuals or regressors of the equation.
Use Tukey fences, mean/standard deviation fences, wavelet outliers, ARMA outliers or influence statistic detection methods to identify observations that may contain outliers.
Syntax
equation_name.resoutliers(options)
Options
sens=arg | Set the sensitivity level. Valid arguments are “low”, “medium” (default), and “high”. |
nofence | Do not perform Tukey and mean/standard deviation fences. |
nowave | Do not perform Wavelet Outlier detection. |
noarma | Do not perform ARMA based outlier detection. ARMA outlier detection is only available for least squares equations containing ARMA terms, and is turned on by default. |
noinf | Do not perform influence statistic (not including DFBETAS) based outlier detection. Influence statistic outlier detection is only available for linear least squares equations, and is turned on by default. |
dfbeta | Perform DFBETA influence statistic based outlier detection. DFBETA based outlier detection is only available for linear least squares equations, and is turned off by default. |
tukeyk=arg | Set the value k in the Tukey fence detection routine. This will override the value of k set by the sens= option. |
meanstdevk=arg | Set the value k in the mean/standard deviation fence detection routine. This will override the value of k set by the sens= option. |
wavesig=arg | Set the value false discovery rate significance value used in the Wavelet Outlier detection routine. This will override the value set by the sens= option. |
armac=arg | Set the value c in the ARMA outlier detection routine. This will override the value of c set by the sens= option. |
rsbound=arg | Set the value c in RSTUDENT outlier detection. This will override the value of c set by the sens= option. |
hbound=arg | Set the value c in HatMatrix outlier detection. This will override the value of c set by the sens= option. |
dfsbound=arg | Set the value c in DFFITS outlier detection. This will override the value of c set by the sens= option. |
covbound=arg | Set the value c in CovRatio outlier detection. This will override the value of c set by the sens= option. |
betabound=arg | Set the value c in DFBETA outlier detection. This will override the value of c set by the sens= option. |
series=name | Create a new series in the workfile, named name, containing a value of 1 for any observations identified as an outlier, and a value of 0 for any observation identified as not an outlier. |
datestring=name | Create a new string object in the workfile containing the dates (or observation identifiers) for any observations identified as an outlier. |
grlabels | Turn on observation labels on the outlier graph. |
Examples
equation eq01.ls gdpc1 c unemp
eq01.resoutliers(nofence, dfbeta, sens=low)
Estimates an equation with GDPC1 as the dependent variable, and a constant and UNEMP as regressors. Then, outlier detection on the residuals is performed, opting to not use either fence detection, but to include the dfbeta influence statistics (along with the other influence statistics included by default), and setting the sensitivity of the detection to "low".
Displays the results view of an estimated equation.
Syntax
equation_name.results(options)
Options
Examples
equation eq1.ls m1 c inc tb3 ar(1)
eq1.results(p)
estimates an equation using least squares, and displays and prints the results.
Cross-references
Display regime probabilities for a switching regression equation.
Syntax
eq_name.rgmprobs(options) [indices]
where eq_name is the name of an equation estimated using switching regression. The elements to display are given by the optional indices corresponding to the regimes (e.g., “1 2 3” or “2 3”). If indices is not provided, results for all of the regimes will be displayed.
Options
type=arg (default=“pred”) | Type of regime probability to compute: one-step ahead predicted (“pred”), filtered (“filt”), smoothed (“smooth”). |
view=arg (default=“graph”) | Display format: multiple graphs (“graph”), single graph “graph1”, sheet (“sheet”), summary (“summary”). |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eq1.switchreg(type=markov) y c @nv ar(1) ar(2) ar(3)
eq1.rgmprobs
displays two graphs containing the one-step ahead regime probabilities for the Markov switching regression estimated in EQ1.
eq1.rgmprobs(type=filt) 2
displays the filtered probabilities for regime 2.
eq1.rgmprobs(type=smooth, view=graph1)
displays the smoothed probabilities for both regimes in a single graph.
Cross-references
Recursive least squares regression.
The rls view of an equation displays the results of recursive least squares (rolling) regression. This view is only available for (non-panel) equations estimated by ordinary least squares without ARMA terms.
You may plot various statistics from rls by choosing an option.
Syntax
eq_name.rls(options) c(1) c(2) …
Options
r | Plot the recursive residuals about the zero line with plus and minus two standard errors. |
r,s | Plot the recursive residuals and save the residual series and their standard errors as series named R_RES and R_RESSE, respectively. |
c | Plot the recursive coefficient estimates with two standard error bands. |
c,s | Plot the listed recursive coefficients and save all coefficients and their standard errors as series named R_C1, R_C1SE, R_C2, R_C2SE, and so on. |
o | Plot the p-values of recursive one-step Chow forecast tests. |
n | Plot the p-values of recursive n-step Chow forecast tests. |
q | Plot the CUSUM (standardized cumulative recursive residual) and 5 percent critical lines. |
v | Plot the CUSUMSQ (CUSUM of squares) statistic and 5 percent critical lines. |
prompt | Force the dialog to appear from within a program. |
p | Print the view. |
Examples
equation eq1.ls m1 c tb3 gdp
eq1.rls(r,s)
eq1.rls(c) c(2) c(3)
plots and saves the recursive residual series from EQ1 and their standard errors as R_RES and R_RESSE. The third line plots the recursive slope coefficients of EQ1.
equation eq2.ls m1 c pdl(tb3,12,3) pdl(gdp,12,3)
eq2.rls(c) c(3)
eq2.rls(q)
The second command plots the recursive coefficient estimates of PDL02, the linear term in the polynomial of TB3 coefficients. The third line plots the CUSUM test statistic and the 5% critical lines.
Cross-references
Estimates an equation using robust least squares.
You may perform three different types of robust estimation: M-estimation, S-estimation and MM-estimation.
Syntax:
eq_name.robustls(options) y x1 [x2 x3…]
Enter the robustls keyword, followed by the dependent variable and a list of the regressors.
Options
method=arg (default=“m”) | Robust estimation method: “m” (M-estimation), “s” (S-estimation) or “mm” (MM-estimation). |
cov=arg (default=“type1”) | Covariance method type: “type1”, “type2”, or “type3”. |
tuning=number | Specify a value for the tuning parameter. If a value is not specified, EViews will use the default tuning parameter for the type of estimation and weighting function (if applicable). |
c=s | Convergence criterion. The criterion will be set to the nearest value between 1e-24 and 0.2. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
m=integer | Maximum number the number of iterations. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
M-estimation Options
fn=arg (default=“bisquare”) | Weighting function used during M-estimation: “andrews” (Andrews), “bisquare” (Bisquare), “cauchy” (Cauchy), “fair”, “huber”, “huberbi” (Huber-bisquare), “logistic” (Logistic), “median”, “tal” (Talworth), “Welsch” (Welsch). |
scale=arg (default=“madzero”) | Scaling method used for calculating the scalar parameter during M estimation: “madzero” (median absolute deviation, zero centered), “madmed” (median absolute deviation, median centered), "huber" (Huber scaling). |
hmat | Use the hat-matrix to down-weight observations with high leverage. |
S and MM estimation options
compare = integer (default=4) | Number of comparison sets. |
refine = integer (default= 2) | Number of refinements. |
trials = integer (default=200) | Number of trials. |
subsmpl=integer | Specifies the size of the subsamples. Note, the default is number of coefficients in the regression. |
seed=number | Specifies the random number generator seed |
rng=arg | Specifies the type of random number generator. The key can be; improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple, recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
MM estimation options
mtuning=arg | M-estimator tuning parameter. Note the S-estimator tuning parameter is set with the “tuning=” option outlined above. |
hmat | Use the hat-matrix to down-weight observations with high leverage during m-estimation. |
Examples
The following examples use the “Rousseeuw and Leroy.wf1” file located in the EViews application data directory.
equation eq1.robustls salinity c lagsal trend discharge
This line estimates a simple M-type robust estimation, with SALINITY as the dependent variable, and a constant, LAGSAL, TREND and DISCHARGE as independent variables.
The line:
equation eq2.robustls(method=mm, tuning=2.937, mtuning=3.44, cov=type2) salinity c lagsal trend discharge
estimates the same model, but using MM-estimation, with an S tuning constant of 2.937, an M tuning constant of 3.44, and using Huber Type II standard errors.
Cross-references
Set the object attribute.
Syntax
equation_name.setattr(attr) attr_value
Sets the attribute attr to attr_value. Note that quoting the arguments may be required. Once added to an object, the attribute may be extracted using the @attr data member.
Examples
a.setattr(revised) never
String s = a.@attr("revised")
sets the “revised” attribute in the object A to the string “never”, and extracts the attribute into the string object S.
Cross-references
Set or clear the local pilot bandwidth. Once set, the cached value may be used in all post-estimation routines that require a pilot bandwidth.
For equations estimated using the functional coefficients method.
Syntax
eq_name.setpilotbw(options)
Options
Basic Options
clear | Clear any previously set local pilot bandwidth.s |
Pilot Bandwidth Options
Unless clearing the local pilot bandwidth using the option “clear”, the following options specify the pilot bandwidth computation.
plth =arg (default = “cv”) | Pilot bandwidth method: simple rule-of-thumb (“rot”), robust rule-of-thumb (“rotr”), residual squares criterion (“rsc”), modified multi cross-validation (“cv”), user-defined (“user”). |
pltbw=arg (default =1) | User-defined bandwidth (if “plth=user”). |
plthmin=arg (default = 0.1) | Bandwidth grid search minimum value (if not “plth=user”). |
plthmax=arg (default =1) | Bandwidth grid search maximum value (if not “plth=user”). |
plthlen=integer (default = 100) | Bandwidth grid search length (if not “plth=user”). |
plthinc=integer (default = 10) | Bandwidth grid search increment step percentage increase (if not “plth=user”). |
plthcup=integer (default = 10) | Stop rule: consecutive increases of objective function before stop (not available when “plth=user”). |
pltm=arg (default = 10) | Modified multifold CV m-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
pltq=integer (default = 4) | Modified multifold CV Q-value: percentage of sample size used in bandwidth determination (when “plth=cv”). |
auxk=integer (default = 2) | Estimation polynomial degree for pilot stage in excess of final stage degree. This number should always be an even positive integer. |
Examples
eq1.setpilotbw.
sets the local pilot bandwith using computation default options.
eq1.setpilotbw(plth=rsc)
computes a pilot bandwidth using the residual squares criterion and saves the value as the local pilot bandwidth.
eq1.setpilotbw(plth=user, pltbw=0.5)
sets the local pilot bandwidth to 0.5.
eq1.setpilotbw(clear)
clears (uninitializes) the local pilot bandwidth.
Cross-references
See
“Functional Coefficient Regression” for discussion of functional coefficients estimation.
See
“Bandwidth Selection” and
“Bandwidth Views” for a discussion of bandwidths.
Perform the sign-bias test (Engle and Ng, 1993) of misspecification in equations estimated using GARCH.
Syntax
eq_name.signbias(options)
Options
p | Print output from the test. |
Examples
equation eq1.arch(2, 1) y c
estimates a GARCH(2, 1) model.
eq1.signbias
displays the results of a sign-bias misspecification test.
Cross-references
See
“ARCH and GARCH Estimation” for a discussion of ARCH models.
Compute the PMG Hausman test for similarity against mean-group and dynamic fixed effects estimators (in panel equations estimated by ARDL/PMG)
Syntax
eq_name.similarity(options)
Options
Example
pmg_eq.similarity
displays a spool object with several tables containing the results of the Hausman test, comparisons of results, and auxiliary estimation results employed in computing the test statistic.
Cross-references
Displays a spool object with the results of error-correction regressions for each cross-section in PMG estimation.
Syntax
eq_name.srcoefs
Options
Example
equation eq.ardl log(cons) log(inf)
eq.srcoefs
Displays a spool object with the results of error-correction regressions for each cross-section in PMG estimation.
Cross-references
Compute tests of parameter constancy of the base specification against a smooth transition alternative in a smooth threshold regression.
Syntax
eq_name.strconstant(options)
Options
p | Print output from the test. |
Cross-references
Compute tests for linearity of the base specification against the smooth threshold alternative in a smooth threshold regression.
Syntax
eq_name.strlinear(options)
Options
p | Print output from the test. |
Cross-references
Compute tests for additional nonlinearity against additive or encapsulated alternatives (for equations in a smooth threshold regression).
Syntax
eq_name.strnonlin(options)
Options
encap | Compute tests for additional nonlinearity against encapsulated alternatives. |
p | Print output from the test. |
Cross-references
Compute and display the transition weights in a smooth threshold regression.
The default display shows a graph of the transition function. You may also display the weights for each observation in the estimation sample.
Syntax
eq_name.strwgts(options)
Options
view=arg | Weight display: “graph” (graph of the weight for each individual in the estimation sample), “sheet” (spreadsheet containing weights for each individual), “summary” (summary statistics). The default view displays a graph of the function with optional borders. |
ab=arg | Additional graph borders to display when showing the default view of the weights: “none” (do not display borders)”, “boxplot” (display boxplot borders), “histogram” (display a histogram). The default view shows a boxplot on each border. |
output=arg | Optional name of matrix to save the data used in the function plot. |
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Cross-references
Estimate a switching regression model (simple exogenous or Markov).
Syntax
eq_name.switchreg(options) dependent_var list_of_varying_regressors [ @nv list_of_nonvarying_regressors ] [ @prv list_of_probability_regressors ]
List the switchreg keyword, followed by options, then the dependent variable and a list of the regressors with regime-varying coefficients, following optionally by the keyword @nv and a list of regressors with regime-invariant coefficients, and by the keyword @prv and a list of regressors that enter into the transition probability specification.
The dependent variable in switchreg may not be an expression. Dynamics may be specified by including lags of the dependent variable as regressors, or by specifying AR errors using the AR keyword. The latter incorporate mean adjusted lags of the form specified by the “Hamilton-model.”
Options
type=arg | Type of switching: simple exogenous (“simple”), Markov (“markov”). |
nstates=integer (default=2) | Number of regimes. |
heterr | Allow for heterogeneous error variances across regimes |
fprobmat=arg | Name of fixed transition probability matrix allows for fixing specific elements of the time-invariant transition matrix. Leave NAs in elements of the matrix to estimate. The 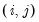 element of the matrix corresponds to 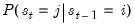 . |
initprob=arg (default=“ergodic”) | Method for determining initial Markov regime probabilities: ergodic solution (“ergodic”), estimated parameter (“est”), equal probabilities (“uniform”), user-specified probabilities (“user”). If “initprob=user” is specified, you will need to specify the “userinit=” option. |
userinit=arg | Name of vector containing user-specified initial Markov probabilities. The vector should have rows equal to the number of states; we expand this to the size of the initial lag state vector where necessary for AR specifications. For use in specifications containing both the “type=markov” and “initprob=user” options. |
startnum=arg (default=0 or 25) | Number of random starting values tried. The default is 0 for user-supplied coefficients (option “s”) and 25 in all other cases. |
startiter=arg (default=10) | Number of iterations taken after each random start before comparing objective to determine final starting value. |
searchnum=arg (default=0) | Number of post-estimation perturbed starting values tried. |
searchsds=arg (default=1) | Number of standard deviations to use in perturbed starts (if “searchnum=”) is specified. |
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
rnd= arg (default=“kn” or method previously set using
rndseed | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
In addition to the specification options, there are options for estimation and covariance calculation.
Additional Options
optmethod = arg | Optimization method: “bfgs” (BFGS); “newton” (Newton-Raphson), “opg” or “bhhh” (OPG or BHHH), “legacy” (EViews legacy). BFGS is the default method. |
optstep = arg | Step method: “marquardt” (Marquardt); “dogleg” (Dogleg); “linesearch” (Line search). Marquardt is the default method. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
cov=arg | Covariance method: “ordinary” (default method based on inverse of the estimated information matrix), “huber” or “white” (Huber-White sandwich method). |
covinfo = arg | Information matrix method: “opg” (OPG); “hessian” (observed Hessian). (Applicable when non-legacy “optmethod=”.) |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
s | Use the current coefficient values in “C” as starting values (see also
param). |
s=number | Specify a number between zero and one to determine starting values as a fraction of EViews default values (out of range values are set to “s=1”). |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eq_41a.switchreg(type=markov) y c @nv ar(1) ar(2) ar(3) ar(4)
estimates a Hamilton-type Markov switching regression model with four non-regime varying autoregressive terms implying mean adjustment for the lagged endogenous.
equation eq_lagdep.switchreg(type=markov) y c @nv y(-1) y(-2) y(-3) y(-4)
specifies an alternate dynamic model in which the lags enter directly into the contemporaneous equation without mean adjustment.
equation eq_filardo.switchreg(type=markov) yy_dalt c @nv ar(1) ar(2) ar(3) ar(4) @prv c yy_ldalt
estimates a 2 state model with non-varying AR(4) and transition matrix probability regressor YY_LDALT.
Cross-references
See
“Switching Regression” for a description of the switching regression methodology.
See also
Equation::rgmprobs,
Equation::transprobs,
Equation::makergmprobs and
Equation::maketransprobs for routines that allow you to work with the regime probabilities and transition probabilities.
Compute symmetry test for distributed lag variables in an equation estimated with a nonlinear ARDL (NARDL) specification.
This view displays a table object with the NARDL symmetry test. The top part of the table is a is a summary of the test. This is followed by three additional sections with test statistics and corresponding p-values for relevant regressors tests for: 1) long-run asymmetry, 2) short-run asymmetry, 3) both long and short-run asymmetry.
Syntax
eq_name.symmtest(options)
Options
Example
ardl_eq.symmtest
computes the NARDL symmetry tests for relevant regressors.
Cross-references
See
“Symmetry Test View” for further discussion.
Test whether to add regressors to an estimated equation.
Tests the hypothesis that the listed variables were incorrectly omitted from an estimated equation (only available for equations estimated by list). The test displays some combination of Wald and LR test statistics, as well as the auxiliary regression.
Syntax
eq_name.testadd(options) arg1 [arg2 arg3 ...]
eq_name.testadd(options) arg1 [arg2 arg3 ...] [@nv x1 x2 x3 ...]
List the names of the series or groups of series to test for omission after the keyword.
For equations estimated using breakls, there are two types of added series, those with coefficients that break, and those with coefficients that are non-breaking. The former should be listed before, and the latter should be listed after the optional keyword.
Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
equation oldeq.ls sales c adver lsales ar(1)
oldeq.testadd gdp gdp(-1)
tests whether GDP and GDP(-1) belong in the specification for SALES using the equation OLDEQ.
Cross-references
See
“Coefficient Diagnostics” for further discussion.
Test whether to drop regressors from a regression.
Tests the hypothesis that the listed variables were incorrectly included in the estimated equation (only available for equations estimated by list). The test displays some combination of
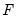
and LR test statistics, as well as the test regression.
Syntax
eq_name.testdrop(options) arg1 [arg2 arg3 ...]
List the names of the series or groups of series to test for omission after the keyword.
Options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
equation oldeq.ls sales c adver lsales ar(1)
oldeq.testdrop adver
tests whether ADVER should be excluded from the specification for SALES using a the equation OLDEQ.
Cross-references
See
“Coefficient Diagnostics” for further discussion of testing coefficients.
Carry out the Hosmer-Lemeshow and/or Andrews goodness-of-fit tests for estimated binary models.
Syntax
binary_equation.testfit(options)
Options
h | Group by the predicted values of the estimated equation. |
s=series_name | Group by the specified series. |
integer (default=10) | Specify the number of quantile groups in which to classify observations. |
u | Unbalanced grouping. Default is to randomize ties to balance the number of observations in each group. |
v | Group according to the values of the reference series. |
l=integer (default=100) | Limit the number of values to use for grouping. Should be used with the “v” option. |
prompt | Force the dialog to appear from within a program. |
p | Print the result of the test. |
Examples
equation eq1.binary work c age edu
eq1.testfit(h,5,u)
estimates a probit specification, and tests goodness-of-fit by comparing five unbalanced groups of actual data to those estimated by the model.
Cross-references
See
“Goodness-of-Fit Tests” for a discussion of the Andrews and Hosmer-Lemeshow tests.
Estimation by discrete or smooth threshold least squares, including threshold autoregression.
Syntax
eq_name.threshold(options) y z1 [z2 z3 ...] [@nv x1 x2 x3 ...] @thresh t1 [t2 t3 ...]
List the dependent variable first, followed by a list of the independent variables that have coefficients that are allowed to vary across threshold, followed optionally by the keyword @nv and a list of non-varying coefficient variables.
List a threshold variable or variables (for model selection) or a single integer or range pairs after the keyword @thresh. The integer or range pairs indicate a self-exciting model with the lagged dependent variable as the threshold variable.
For smooth threshold equations you may specify variables that are to be included only in the base specification or only in the alternative specification. Base-only variables should be specified in parentheses using the @base key, as in “@base(x1) @base(x2) @base(x3 x4)”. Alternative-only variables may be specified analogously using the @alt key.
Options
Specification Options
type=arg (default=“discrete”) | Type of threshold estimation: “discrete” (discrete), “smooth” (smooth). |
Discrete Threshold Options
method=arg (default=“seqplus1”) | Threshold selection method: “seqplus1” (sequential tests of single 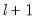 versus 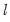 thresholds), “seqall” (sequential test of all possible 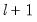 versus 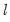 thresholds), “glob” (tests of global 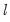 vs. no thresholds), “globplus1” (tests of 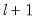 versus 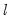 globally determined thresholds), “globinfo” (information criteria evaluation)., “fixedseq” (fixed number of sequentially determined thresholds), “fixedglob” (fixed number of globally determined thresholds), “user” (user-specified thresholds) |
nthresh=arg (default=1) | Number of thresholds for fixed number threshold selection methods. |
select=arg | Sub-method setting (options depend on “method=”). (1) if “method=glob”: Sequential ("seq") (default), Highest significant ("high"), 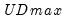 ("udmax"), 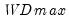 ("wdmax"). (2) if “method=globinfo”: Schwarz criterion (“bic” or “sic”) (default), Liu-Wu-Zidek criterion (“lwz”). |
trim=arg (default=5) | Trimming percentage for determining minimum segment size (5, 10, 15, 20, 25). |
maxthresh=integer (default=5) | Maximum number of thresholds to allow (not applicable if “method=seqall”). |
maxlevels=integer (default=5) | Maximum number of threshold levels to consider in sequential testing (applicable when “method=sequall”). |
size=arg (default=5) | Test sizes for use in sequential determination and final test evaluation (10, 5, 2.5, 1) corresponding to 0.10, 0.05, 0.025, 0.01, respectively |
heterr | Assume regimes specific error distributions in variance computation. |
commondata | Assume a common distribution for the data across segments (only applicable if original equation is estimated with a robust covariance method, “heterr” is not specified). |
Smooth Threshold Options
smoothtrans=arg (default=“logistic”) | Smooth threshold transition function: “logistic” (logistic), “logistic2” (second-order logistic), “exponential” (exponential), “normal” (normal). |
smoothstart=arg (default=“grid_conc”) | Smoth threshold starting value method: or fixed number threshold selection methods: “grid_conc” (grid search with concentrated regression coefficients”, “grid_zeros” (grid search with zero regression coefficients), “data” (data-based), “user” (user-specified using the contents of the coefficient vector in the workfile). |
smoothst=arg | Sub-method setting (options depend on “method=”). (1) if “method=glob”: Sequential ("seq") (default), Highest significant ("high"), 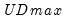 ("udmax"), 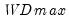 ("wdmax"). (2) if “method=globinfo”: Schwarz criterion (“bic” or “sic”) (default), Liu-Wu-Zidek criterion (“lwz”). |
General Options
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=keyword | Covariance type (optional): “white” (White diagonal matrix), “hac” (Newey-West HAC). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 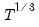 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Examples
equation eq1.threshold(method=fixedseq, type=discrete) ss_transf c ss_transf(-1 to -11) @thresh 2
uses the fixed number of thresholds test to determine the optimal threshold in a model regressing SS_TRANSF on the threshold variables C and SS_TRANSF(-1 to -11).
equation eq2.threshold(method=fixedseq, type=discrete) ss_transf c ss_transf(-1 to -11) @thresh 1 5
uses the fixed number of thresholds test to determine the optimal threshold and does model selection over lags of SS_TRANSF from SS_TRANSF(-1) to SS_TRANSF(-5).
equation eq3.threshold(method=user, threshold=7.44) ss_transf c @nv ss_transf(-1 to -11) @thresh 2
estimates the model with one user-specified threshold value. In addition, the variables SS_TRANSF(-1 to -11) are restricted to have common coefficients across the regimes.
Cross-references
See
“Discrete Threshold Regression” and
“Smooth Transition Regression” for a discussion of the various forms of threshold models.
Display regime transition probabilities and expected durations for a switching regression equation.
Syntax
equation_name.transprobs(options)
where equation_name is the name of an equation estimated using switching regression.
Options
type=arg (default=“summary”) | Transition probability results to display: summary (“default”), transition probabilities (“trans”), expected durations (“expect”). The default summary displays the transition matrix and expected regime durations for constant transition probability models, and descriptive statistics for the transition and expected durations for varying probability models. |
view=arg (default=“graph”) | Display method: graph (“graph”), spreadsheet (“sheet”), table (“table”). Applicable when displaying the transition probabilities or expected durations (“type=trans” or “type=expect”). The spreadsheet form represents shows the transition probabilities or regime expected durations in columns and observations in rows. The table form displays the transition probabilities or expected durations in a table (in a single matrix for a time-constant model, and individual matrices for a time-varying model). |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
equation eq1.switchreg(type=markov) y c @nv ar(1) ar(2) ar(3)
eq1.transprobs
displays the default summary of the transition probabilities estimated in EQ1.
The command
eq1.transprobs(type=trans)
displays the transition probabilities in a graph, while
eq1.transprobs(type=trans, view=sheet)
displays the transition probabilities in a spreadsheet, with each row column representing one of the probabilities and each row representing an observation.
eq1.transprobs(type=trans, view=table)
displays the transition probabilities in a table.
eq1.transprobs(type=expect, view=sheet)
displays the expected durations in spreadsheet form.
Cross-references
Two-stage least squares.
Carries out estimation for equations using two-stage least squares.
Syntax
eq_name.tsls(options) y x1 [x2 x3 ...] @ z1 [z2 z3 ...]
eq_name.tsls(options) specification @ z1 [z2 z3 ...]
To use the tsls command, list the dependent variable first, followed by the regressors, then any AR or MA error specifications, then an “@”-sign, and finally, a list of exogenous instruments. You may estimate nonlinear equations or equations specified with formulas by first providing a specification, then listing the instrumental variables after an “@”-sign.
There must be at least as many instrumental variables as there are independent variables. All exogenous variables included in the regressor list should also be included in the instrument list. A constant is included in the list of instrumental variables even if not explicitly specified.
Options
Non-Panel TSLS Options
nocinst | Do not automatically include a constant as an instrument. |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
cov=keyword | Covariance type (optional): “white” (White diagonal matrix), “hac” (Newey-West HAC), “cr” (cluster robust). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. (For non-cluster robust methods). |
covlag=arg (default=1) | Whitening lag specification: integer (user-specified lag value), “a” (automatic selection). |
covinfosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
covmaxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 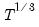 . |
covkern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
covbw=arg (default=“fixednw”) | Kernel Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
covnwlag=integer | Newey-West lag-selection parameter for use in nonparametric kernel bandwidth selection (if “covbw=neweywest”). |
covbwint | Use integer portion of bandwidth. |
crtype=arg (default “cr1”) | Cluster robust weighting method: “cr0” (no finite sample correction), “cr1” (finite sample correction), when “cov=cr”. |
crname=arg | Cluster robust series name, when “cov=cr”. |
m=integer | Set maximum number of iterations. |
c=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR or MA terms (see also
param). |
s=number | Determine starting values for equations specified by list with AR or MA terms. Specify a number between zero and one representing the fraction of TSLS estimates computed without AR or MA terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR and MA terms which are instead set to EViews determined default values. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
z | Turn off backcasting in ARMA models. |
prompt | Force the dialog to appear from within a program. |
p | Print basic estimation results. |
Panel TSLS Options
cx=arg | Cross-section effects. For fixed effects estimation, use “cx=f”; for random effects estimation, use “cx=r”. |
per=arg | Period effects. For fixed effects estimation, use “cx=f”; for random effects estimation, use “cx=r”. |
wgt=arg | GLS weighting: (default) none, cross-section system weights (“wgt=cxsur”), period system weights (“wgt=persur”), cross-section diagonal weighs (“wgt=cxdiag”), period diagonal weights (“wgt=perdiag”). |
cov=arg | Coefficient covariance method: (default) ordinary, White cross-section system robust (“cov=cxwhite”), White period system robust (“cov=perwhite”), White heteroskedasticity robust (“cov=stackedwhite”), Cross-section system robust/PCSE (“cov=cxsur”), Period system robust/PCSE (“cov=persur”), Cross-section heteroskedasticity robust/PCSE (“cov=cxdiag”), Period heteroskedasticity robust (“cov=perdiag”). |
keepwgts | Keep full set of GLS weights used in estimation with object, if applicable (by default, only small memory weights are saved). |
rancalc=arg (default=“sa”) | Random component method: Swamy-Arora (“rancalc=sa”), Wansbeek-Kapteyn (“rancalc=wk”), Wallace-Hussain (“rancalc=wh”). |
nodf | Do not perform degree of freedom corrections in computing coefficient covariance matrix. The default is to use degree of freedom corrections. |
iter=arg (default=“onec”) | Iteration control for GLS specifications: perform one weight iteration, then iterate coefficients to convergence (“iter=onec”), iterate weights and coefficients simultaneously to convergence (“iter=sim”), iterate weights and coefficients sequentially to convergence (“iter=seq”), perform one weight iteration, then one coefficient step (“iter=oneb”). Note that random effects models currently do not permit weight iteration to convergence. |
unbalsur | Compute SUR factorization in unbalanced data using the subset of available observations for a cluster. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default is to use the “C” coefficient vector. |
s | Use the current coefficient values in estimator coefficient vector as starting values for equations specified by list with AR terms (see also
param). |
s=number | Determine starting values for equations specified by list with AR terms. Specify a number between zero and one representing the fraction of TSLS estimates computed without AR terms to be used. Note that out of range values are set to “s=1”. Specifying “s=0” initializes coefficients to zero. By default EViews uses “s=1”. Does not apply to coefficients for AR terms which are instead set to EViews determined default values. |
m=integer | Set maximum number of iterations. |
c=number | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled coefficients. The criterion will be set to the nearest value between 1e-24 and 0.2. |
numericderiv / ‑numericderiv | [Do / do not] use numeric derivatives only. If omitted, EViews will follow the global default. |
fastderiv / ‑fastderiv | [Do / do not] use fast derivative computation. If omitted, EViews will follow the global default. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the estimation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
eq1.tsls y_d c cpi inc ar(1) @ lw(-1 to -3)
estimates EQ1 using TSLS regression of Y_D on a constant, CPI, INC with AR(1) using a constant, LW(-1), LW(-2), and LW(-3) as instruments.
param c(1) .1 c(2) .1
eq1.tsls(s,m=500) y_d=c(1)+inc^c(2) @ cpi
estimates a nonlinear TSLS model using a constant and CPI as instruments. The first line sets the starting values for the nonlinear iteration algorithm.
Cross-references
See
“Instrumental Variables and GMM” and
“Two-Stage Least Squares” for details on two-stage least squares estimation in single equations and systems, respectively.
“Instrumental Variables” discusses estimation using pool objects, while
“Instrumental Variables Estimation” discusses estimation in panel structured workfiles.
Andrews-Quandt test for unknown breakpoint.
Carries out the Andrews-Quandt test for parameter stability at some unknown breakpoint.
Syntax
eq_name.ubreak(options) trimlevel @ x1 x2 x3
You must provide the level of trimming of the data. The level must be one of the following: 49, 48, 47, 45, 40, 35, 30, 25, 20, 15, 10, or 5. If the equation is specified by list and contains no nonlinear terms, you may specify a subset of the regressors to be tested for a breakpoint after an “@” sign.
Options
wfname = series_name | Store the individual Wald F-statistics into the series series_name. |
lfname = series_name | Store the individual likelihood ratio F-statistics into the series series_name. |
prompt | Force the dialog to appear from within a program. |
p | Print the result of the test. |
Examples
equation ppp.ls log(spot) c log(p_us) log(p_uk)
ppp.ubreak 15
regresses the log of SPOT on a constant, the log of P_US, and the log of P_UK, and then carries out the Andrews-Quandt test, trimming 15% of the data from each end.
To test whether only the constant term and the coefficient on the log of P_US are subject to a structural break, use:
ppp.ubreak @ c log(p_us)
Cross-references
Update coefficient object values from an equation object.
Copies coefficients from the equation object into the appropriate coefficient vector or vectors.
Syntax
equation_name.updatecoef
Follow the name of the equation object with a period and the keyword updatecoef.
Examples
equation eq1.ls y c x1 x2 x3
equation eq2.ls z c z1 z2 z3
eq1.updatecoef
places the coefficients from EQ1 in the default coefficient vector C.
coef(3) a
equation eq3.ls y=a(1)+z1^c(1)+log(z2+a(2))+exp(c(4)+z3/a(3))
equation eq2.ls z c z1 z2 z3
eq3.updatecoef
updates the coefficient vector A and the default vector C so that both contain the coefficients from EQ3.
Cross-references
Variance Inflation Factor (VIF).
Display the Variance Inflation Factors (VIFs). VIFs are a method of measuring the level of collinearity between the regressors in an equation.
Syntax
eq_name.varinf
Options
Examples
The set of commands:
equation eq1.ls lwage c edu edu^2 union
eq1.varinf
displays the variance inflation factor view of EQ1.
Cross-references
Estimation using variable selection.
Syntax
eq_name.varsel(options) y x1 [x2 x3 ...] @ z1 z2 z3
Specify the dependent variable followed by a list of variables to be included in the regression, but not part of the search routine, followed by an “@” symbol and a list of variables to be part of the search routine. If no included variables are required, simply follow the dependent variable with an “@” symbol and the list of search variables.
Options
method = arg | Stepwise regression method: “stepwise” (default), “uni” (uni-directional), “swap” (swapwise), “comb” (combinatorial), “gets” (auto-search/GETS), “lasso” (Lasso). |
nvars = int | Set the number of search regressors. Required for swapwise and combinatorial methods, optional for uni-directional and stepwise methods. |
w=arg | Weight series or expression. Note: we recommend that, absent a good reason, you employ the default settings (“wtype=istdev”) with scaling (“wscale=eviews”) for backward compatibility with versions prior to EViews 7. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Stepwise and uni-directional method options
back | Set stepwise or uni-directional method to run backward. If omitted, the method runs forward. |
tstat | Use t-statistic values as a stopping criterion. (default uses p-values). |
ftol=number (default = 0.5) | Set forward stopping criterion value. |
btol=number (default = 0.5) | Set backward stopping criterion value. |
fmaxstep=int (default = 1000) | Set the maximum number of steps forward. |
bmaxstep=int (default = 1000) | Set the maximum number of steps backward. |
tmaxstep=int (default = 2000) | Set the maximum total number of steps. |
Swapwise method options
minr2 | Use minimum R-squared increments. (Default uses maximum R-squared increments.) |
Combinatorial method options
force | Suppress the warning message issued when a large number of regressions will be performed. |
Auto-search/GETS method options
pval=number (default = 0.05) | Set the terminal condition p-value used to determine the stopping point of each search path |
nolm | Do not perform AR LM diagnostic test. |
arpval=number (default = 0.025) | Set p-value used in AR LM diagnostic test. |
arlags=int (default = 1) | Set number of lags used in AR LM diagnostic test. |
noarch | Do not perform ARCH LM diagnostic test. |
archpval=number (default = 0.025) | Set p-value used in ARCH LM diagnostic test. |
archlags=int (default = 1) | Set number of lags used in ARCH LM diagnostic test. |
nojb | Do not perform Jarque-Bera normality diagnostic test. |
jbpval=number (default = 0.025) | Set p-value used in Jarque-Bera normality diagnostic test. |
nopet | Do not perform Parsimonious Encompassing diagnostic test. |
petpval=number (default = 0.025) | Set p-value used in Parsimonious Encompassing diagnostic test. |
nogum | Do not include the general model as a candidate for model selection. |
noempty | Do not include the empty model as a candidate for model selection. |
ic =arg | Set the information criterion used in model selection: “AIC” (Akaike information criteria, default), “BIC” (Schwarz information criteria), “HQ” (Hannan-Quin criteria). |
blocks=int | Override the EViews’ determination of the number of blocks in which to split the estimation sample. |
Lasso method options
Penalty Options
ytrans=arg (default=“none”) | Scaling of the dependent variable: “none” (none), “L1” (L1), “L2” (L2), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
xtrans=arg (default=“stdpop”) | Scaling of the regressor variables: “none” (none), “L1” (L1 norm), “L2” (L2 norm), “stdsmpl” (sample standard deviation), “stdpop” (population standard deviation), “minmax” (min-max). |
lambda=arg | Value of the penalty parameter. Can be a single number, list of space-delimited numbers, a workfile series object, or left blank for a EViews determined path (default). Values must be zero or greater. |
nlambdas=integer (default=100) | Number of penalty values for EViews-supplied list. |
nlambdamin=integer (default=5) | Minimum number of lambda values in the path before applying stopping rules. |
minddev=arg (default=1e-05) | Minimum change in deviance fraction to continue estimation. Truncate path estimation if relative change in deviance is smaller than this value. |
maxedev=arg (default=0.99) | Maximum of deviance explained fraction attained to terminate estimation. Truncate path estimation if fraction of null deviance explained is larger than this value. |
maxvars=arg | Maximum number of regressors in the model. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) reaches this value. |
maxvarsratio=arg | Maximum number of regressors in the model as a fraction of the number of observations. Truncate path estimation if the number of coefficients (including those for non-penalized variables like the intercept) divided by the number of observations reaches this value. |
Cross Validation Options
cvmethod=arg (default=“kfold_cv”) | Cross-validation method: “kfold” (k-fold), “simple” (simple split), “mcarlo” (Monte Carlo), “leavepout” (leave-P-out), “leave1out” (leave-1-out), “rolling” (rolling window), “expanding” (expanding window). |
cvmeasure=arg (default=“mse”) | Cross-validation fit measure: “mse” (mean-squared error), “r2” (R‑squared), “mae” (mean absolute error), “mape” (mean absolute percentage error), “smape” (symmetric mean absolute percentage error). |
cvnfolds=arg (default=5) | Number of folds for K-fold cross-validation. For “cvmethod=kfold”. |
cvftrain=arg (default=0.8) | Proportion of data for split and Monte Carlo methods. For “cvmethod=simple” and “cvmethod=mcarlo”. |
cvnreps=arg (default=1) | Number of Monte Carlo method repetitions. For “cvmethod=mcarlo”. |
cvleaveout=arg (default=2) | Number of data points left out for leave-p-out method. For “cvmethod=leavepout”. |
cvnwindows=arg (default=4) | Number of windows for rolling window cross-validation method. For “cvmethod=rolling”. |
cvinitial=arg (default=12) | Number of initial data points in the training set for expanding cross-validation. For “cvmethod=expanding”. |
cvpregap=arg (default=0) | Number of observations between end of training set and beginning of test set. For “cvmethod=simple”, “cvmethod=rolling” and “cvmethod=expanding”. |
cvhorizon=arg (default=1) | Number of observation in the test set. For “cvmethod=rolling” and “cvmethod=expanding”. |
cvpostgap=arg (default=0) | Number of observations between end of test set and beginning of next training set for rolling window or between end of test set and end of next training set for expanding window. For “cvmethod=rolling” and “cvmethod=expanding” |
Random Number Options
seed=positive_integer from 0 to 2,147,483,647 | Seed the random number generator. If not specified, EViews will seed random number generator with a single integer draw from the default global random number generator. |
rnd= arg (default=“kn” or method previously set using
rndseed). | Type of random number generator: improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
Other Options
coefmin= vector_name, number | Vector of individual coefficient minimum values, containing negative or missing values sized to and matching the order of the variables in the specification, or a negative value for the minimum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual minimums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
coefmax= vector_name, number | Vector of individual coefficient maximum values, containing positive or missing values sized to and matching the order of the variables in the specification, or a positive value for the maximum for all coefficients. Missing values in the vector should be used to indicate that the coefficient is unrestricted. If a vector of values is provided and individual maximums are specified using one or more @vw regressors, the vector values will be applied first, then overwritten by the individual values. |
maxit=integer | Maximum number of iterations. |
conv=scalar | Set convergence criterion. The criterion is based upon the maximum of the percentage changes in the scaled estimates. The criterion will be set to the nearest value between 1e-24 and 0.2. |
w=arg | Weight series or expression. |
wtype=arg (default=“istdev”) | Weight specification type: inverse standard deviation (“istdev”), inverse variance (“ivar”), standard deviation (“stdev”), variance (“var”). |
wscale=arg | Weight scaling: EViews default (“eviews”), average (“avg”), none (“none”). The default setting depends upon the weight type: “eviews” if “wtype=istdev”, “avg” for all others. |
coef=arg | Specify the name of the coefficient vector (if specified by list); the default behavior is to use the “C” coefficient vector. |
showopts / ‑showopts | [Do / do not] display the starting coefficient values and estimation options in the rotation output. |
prompt | Force the dialog to appear from within a program. |
p | Print estimation results. |
Examples
eq1.varsel(method=comb,nvars=3) y c @ x1 x2 x3 x4 x5 x6 x7 x8
performs a combinatorial search routine to search for the three variables from the set of X1, X2, ..., X8, yielding the largest R-squared in a regression of Y on a constant and those three variables.
Cross-references
Wald coefficient restriction test.
The wald view carries out a Wald test of coefficient restrictions for an equation object.
Syntax
equation_name.wald restrictions
Enter the equation name, followed by a period, and the keyword. You must provide a list of the coefficient restrictions, with joint (multiple) coefficient restrictions separated by commas.
Options
Examples
eq1.wald c(2)=0, c(3)=0
tests the null hypothesis that the second and third coefficients in equation EQ1 are jointly zero.
eq2.wald c(2)=c(3)*c(4)
tests the non-linear restriction that the second coefficient in equation EQ2 is equal to the product of the third and fourth coefficients.
Cross-references
Displays the Weak Instruments Summary
The weakinst view of an equation displays the Weak Instrument Summary for equations estimated by TSLS, GMM or LIML. The summary includes both the Cragg-Donald test and Moment Selection Criteria (for TSLS and GMM only).
Syntax
eq_name.weakinst
Examples
equation eq1.gmm y c x1 x2 @ z1 z2 z3 z4
e1.weakinst
estimates and equation via GMM and then displays the weak instrument summary.
Cross-references
Performs White’s test for heteroskedasticity of residuals.
Carries out White’s test for heteroskedasticity of the residuals of the specified equation. By default, the test is computed without the cross-product terms (using only the terms involving the original variables and squares of the original variables). You may elect to compute the original form of the White test that includes the cross-products.
White’s test is not available for equations estimated by binary, ordered, censored, or count.
Note that a more general version of the White test is available using
Equation::hettest. We also note that for equations estimated without a constant term, version 6 of the White command will, by default, generate results that differ from version 5. You may obtain version 5 compatible results by adding the @
comp keyword to
white as in:
eq_name.white @comp
Syntax
eq_name.white(options)
Options
c | Include all possible nonredundant cross-product terms in the test regression. |
prompt | Force the dialog to appear from within a program. |
p | Print the test results. |
Examples
eq1.white(c)
carries out the White test of heteroskedasticity including all possible cross-product terms.
Cross-references
See
“White's Heteroskedasticity Test” for a discussion of White’s test. For the multivariate version of this test, see
“White Heteroskedasticity Test”.
See also
Equation::hettest for a more full-featured version of this test.
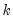 (if applicable).
(if applicable).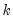 (if applicable).
(if applicable).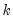 (if applicable).
(if applicable).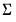 for cointegrating regression residuals estimated with CCR.
for cointegrating regression residuals estimated with CCR.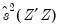 for 2SLS and
for 2SLS and 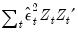 for White weighting).
for White weighting).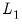 portion of the objective,
portion of the objective, 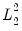 portion of the objective).
portion of the objective).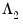 containing the long run covariances of
containing the long run covariances of 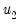 with
with 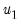 and
and 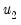 for cointegrating regression equations estimated with CCR and FMOLS.
for cointegrating regression equations estimated with CCR and FMOLS. for cointegrating regression equations estimated with CCR and FMOLS.
for cointegrating regression equations estimated with CCR and FMOLS.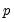 or
or 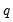 is 9; values above will be set to 9. The minimum value for
is 9; values above will be set to 9. The minimum value for 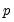 is 1. The minimum value for
is 1. The minimum value for 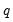 is 0. If either
is 0. If either 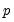 or
or 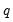 is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default.
is not specified, EViews will assume a corresponding order of 1. Thus, a GARCH(1, 1) is assumed by default. 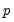 and
and  options should not be specified, and if provided, will be ignored.
options should not be specified, and if provided, will be ignored.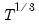 .
.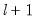 versus
versus 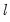 breaks), “seqall” (sequential test of all possible
breaks), “seqall” (sequential test of all possible 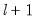 versus
versus 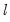 breaks), “glob” (tests of global
breaks), “glob” (tests of global 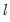 vs. no breaks), “globplus1” (tests of
vs. no breaks), “globplus1” (tests of 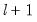 versus
versus 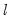 globally determined breaks), “globinfo” (information criteria evaluation),“user” (user-specified break dates).
globally determined breaks), “globinfo” (information criteria evaluation),“user” (user-specified break dates).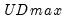 (“udmax”),
(“udmax”), 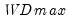 (“wdmax”).
(“wdmax”).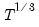 .
.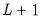 versus
versus 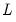 tests to determine the optimal breaks in a model regressing M1 on the breaking variables C and UNEMP.
tests to determine the optimal breaks in a model regressing M1 on the breaking variables C and UNEMP.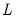 versus none tests to determine the breaks. The selected break will be the highest significant number of breaks.
versus none tests to determine the breaks. The selected break will be the highest significant number of breaks.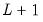 versus
versus 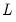 tests to determine the optimal breaks.
tests to determine the optimal breaks.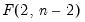 distribution is used; if the parameter estimates are likelihood based, the
distribution is used; if the parameter estimates are likelihood based, the 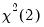 distribution will be employed. “dist=f” forces use of the F-distribution, while “dist=c” uses the
distribution will be employed. “dist=f” forces use of the F-distribution, while “dist=c” uses the 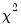 distribution.
distribution.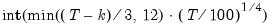
 is the number of coefficients in the cointegrating equation. Applicable when “lag=a”.
is the number of coefficients in the cointegrating equation. Applicable when “lag=a”. , or to place inequality restrictions on the coefficient values, you may do so using special expressions of the form:
, or to place inequality restrictions on the coefficient values, you may do so using special expressions of the form: .
.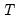 and the number of potential regressors
and the number of potential regressors 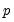 .
.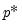 where
where 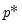 the number of non-zero
the number of non-zero 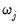 .
. 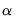 equal to the default of 0.9, no regressor or dependent variable scaling, automatically determined 100-element lambda path with minimum lambda of 0.0001 times the maximum value, using the default K-fold cross-validation with 5 folds with an MSE objective and a random generator seed of 513255899 to determine the optimal value.
equal to the default of 0.9, no regressor or dependent variable scaling, automatically determined 100-element lambda path with minimum lambda of 0.0001 times the maximum value, using the default K-fold cross-validation with 5 folds with an MSE objective and a random generator seed of 513255899 to determine the optimal value.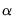 equal to the 0.75 using rolling window cross-validation and SMAPE cross-validation.
equal to the 0.75 using rolling window cross-validation and SMAPE cross-validation.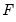 and LR test statistics. If the estimated specification involves two-way fixed effects, three separate tests will be performed; one for each set of effects, and one for the joint effects.
and LR test statistics. If the estimated specification involves two-way fixed effects, three separate tests will be performed; one for each set of effects, and one for the joint effects.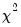 statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”).
statistic (“pearson”), deviance statistic (“deviance”), unit (“unit”), user-specified (“user”).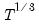 .
.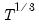 .
.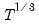 .
.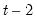 to 1.
to 1.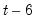 to
to 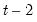 , and ordinary instruments X(-1) and Y(-1).
, and ordinary instruments X(-1) and Y(-1). 
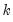 in the K‑class estimator. If omitted, LIML is performed, and
in the K‑class estimator. If omitted, LIML is performed, and 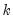 is calculated as part of the estimation procedure.
is calculated as part of the estimation procedure.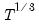 .
.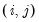 . The series names for expected durations will be of the form base_name# where # corresponds to the regime index. Thus, in a two-regime model, the base name “TEMP” corresponds to the transition probability series TEMP11, TEMP12, TEMP21, TEMP22, and the expected duration series TEMP1, TEMP2.
. The series names for expected durations will be of the form base_name# where # corresponds to the regime index. Thus, in a two-regime model, the base name “TEMP” corresponds to the transition probability series TEMP11, TEMP12, TEMP21, TEMP22, and the expected duration series TEMP1, TEMP2. 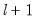 versus
versus 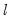 breaks), “seqall” (sequential test of all possible
breaks), “seqall” (sequential test of all possible 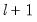 versus
versus 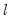 breaks), “glob” (tests of global
breaks), “glob” (tests of global 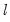 vs. no breaks), “globplus1” (tests of
vs. no breaks), “globplus1” (tests of 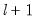 versus
versus 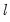 globally determined breaks), “globinfo” (information criteria evaluation).
globally determined breaks), “globinfo” (information criteria evaluation).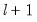 versus
versus 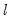 breaks. The second line tests uses the global
breaks. The second line tests uses the global 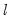 breaks versus none tests with trimming value 0.15, and a size of 0.10 to test for differences in the coefficient on TB3 across regimes.
breaks versus none tests with trimming value 0.15, and a size of 0.10 to test for differences in the coefficient on TB3 across regimes.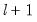 versus
versus  breaks. The third line uses the global
breaks. The third line uses the global  breaks versus none tests. Both of these tests allow for error distributions to vary across the different segments. The final line evaluates the breakpoints using information criteria associated with the global optimizers.
breaks versus none tests. Both of these tests allow for error distributions to vary across the different segments. The final line evaluates the breakpoints using information criteria associated with the global optimizers. 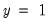 ). Specify the cutoff level as the first option in parentheses after the keyword predict.
). Specify the cutoff level as the first option in parentheses after the keyword predict.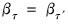 for the slope coefficients
for the slope coefficients 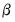 .
.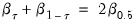 .
.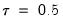 ), EViews will test using estimates obtained at the specified outer quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the outer quartiles {0.25, 0.75}.
), EViews will test using estimates obtained at the specified outer quantiles as specified by the “n=” option. If “n=” is not specified, the default is to display results for the outer quartiles {0.25, 0.75}.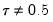 , EViews will include the original quantile in the set of quantiles to test. You may specify “n=e” to perform a test using only estimates obtained at the symmetric pair {
, EViews will include the original quantile in the set of quantiles to test. You may specify “n=e” to perform a test using only estimates obtained at the symmetric pair { ,
,  }.
}.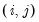 element of the matrix corresponds to
element of the matrix corresponds to  .
. and LR test statistics, as well as the test regression.
and LR test statistics, as well as the test regression. versus
versus 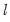 thresholds), “seqall” (sequential test of all possible
thresholds), “seqall” (sequential test of all possible 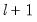 versus
versus 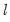 thresholds), “glob” (tests of global
thresholds), “glob” (tests of global 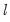 vs. no thresholds), “globplus1” (tests of
vs. no thresholds), “globplus1” (tests of 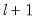 versus
versus 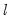 globally determined thresholds), “globinfo” (information criteria evaluation)., “fixedseq” (fixed number of sequentially determined thresholds), “fixedglob” (fixed number of globally determined thresholds), “user” (user-specified thresholds)
globally determined thresholds), “globinfo” (information criteria evaluation)., “fixedseq” (fixed number of sequentially determined thresholds), “fixedglob” (fixed number of globally determined thresholds), “user” (user-specified thresholds) ("udmax"),
("udmax"),  ("wdmax").
("wdmax").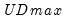 ("udmax"),
("udmax"), 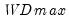 ("wdmax").
("wdmax").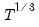 .
.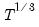 .
.