Group
Group of series. Groups are used for working with collections of series objects (series, alphas, links).
Group Declaration
group create a group object.
To declare a group, enter the keyword group, followed by a name, and optionally, a list of series or expressions:
group salesvrs
group nipa cons(-1) log(inv) g x
You may use the wildcard characters “*” and “?” to match more than one series in the workfile, and you may use the keywords “and” and “not” to specify that certain items should be excluded from the group:
group g a* and *1
makes a group G containing all series whose names begin with the letter “a” and end with “1”, while
group g a* b* not *1 *2
makes a group G containing all series whose names begin with either letter “a” or “b” that do not end with either “1” or “2”.
Additionally, a number of object procedures will automatically create a group.
Group Views
cause pairwise Granger causality tests.
coint test for cointegration between series in a group.
cor correlation matrix between series.
correl correlogram of the first series in the group.
cov covariance matrix between series.
cross cross correlogram of the first two series.
display display table, graph, or spool in object window.
dups duplicates display for observations in the group.
freq frequency table
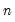
-way contingency table.
label label information for the group.
lrcov compute the symmetric, one-sided, or strict one-sided long-run covariance matrix for a group of series.
members display the members of the group.
pcomp principal components analysis on the members of the group.
sheet spreadsheet view of the series in the group.
stats descriptive statistics.
testbtw tests of equality for mean, median, or variance, between series in group.
uroot independent (panel) unit root test on the series in the group.
uroot2 dependent (second generation panel) unit root tests on the series in the group.
Group Procs
add add one or more series to the group.
clearcontents clear a contiguous block of observations in a group.
clearhist clear the contents of the history attribute.
copy creates a copy of the group.
ddloadtmpl loads a dated data table template for the group.
ddrowopts set the individual row options for the dated data table view of the series in a group.
ddsavetmpl saves the current dated data table settings as a new template.
ddtabopts set the table default options for the dated data table view of the series in a group.
distdata save distribution plot data to a matrix.
drop drop one or more series from the group.
insertobs shift the observations of the group up or downwards, inserting blank observations.
makepcomp save the scores from a principal components analysis of the series in a group.
makesystem creates a system object from the group for other estimation methods.
olepush push updates to OLE linked objects in open applications.
reorder reorder the members of the group.
setattr set the value of an object attribute.
setfillcolor set custom spreadsheet fill coloring for the group.
setformat set the display format in the group spreadsheet for the specified series.
setindent set the indentation in the group spreadsheet for the specified series.
setjust set the justification for cells in the spreadsheet view of the group object.
settextcolor set custom spreadsheet text coloring for the group.
setwidth set the column width in the group spreadsheet for the specified series.
sort change display order for group spreadsheet.
Group Graph Views
Graph creation types are discussed in detail in
“Graph Creation Command Summary”.
area area graph of the series in the group.
bar single or multiple bar graph view of all series.
boxplot boxplot of each series in the group.
hilo high-low(-open-close) chart.
line single or multiple line graph view of all series.
qqplot quantile-quantile plots.
scatmat matrix of all pairwise scatter plots.
Group Data Members
(i) i-th series in the group. Simply append “(i)” to the group name (without a “.”). For use as argument to functions that take a series, not as a series object.
Scalar Values
@comobs number of observations in the current sample for which each series in the group has a non-missing value (observations in the common sample).
@count number of series in the group.
@minobs number of non-missing observations in the current sample for the shortest series in the group.
@maxobs number of non-missing observations in the current sample for the the longest series in the group.
String Values
@attr("arg") string containing the value of the arg attribute, where the argument is specified as a quoted string.
@description string containing the object description (if available).
@depends string containing a list of the series in the current workfile on which this group depends.
@detailedtype string with the object type: “GROUP”.
@displayname string containing the Group’s display name. If the Group has no display name set, the name is returned.
@members string containing a space delimited list of the names of the series contained in the Group.
@name string containing the Group’s name.
@remarks string containing the Group’s remarks (if available).
@seriesname(i) string containing the name of the i-th series in the group.
@type string with the object type: “GROUP”.
@updatetime string representation of the time and date at which the Group was last updated.
Group Examples
To create a group G1, you may enter:
group g1 gdp income
To change the contents of an existing group, you can repeat the declaration, or use the add and drop commands:
group g1 x y
g1.add w z
g1.drop y
The following commands produce a cross-tabulation of the series in the group, display the covariance matrix, and test for equality of variance:
g1.freq
g1.cov
g1.testbtw(var,c)
You can index selected series in the group:
show g1(2).line
series sum=g1(1)+g1(2)
To create a scalar containing the number of series in the group, use the command:
scalar nsers=g1.@count
Add series to a group.
Syntax
group_name.add arg1 [arg2 arg3 ...]
List the names of series or a group of series to add to the group.
Examples
dummy.add d11 d12
Adds the two series D11 and D12 to the group DUMMY.
Cross-references
See
“Groups” for additional discussion of groups.
“Cross-section Identifiers” discusses pool identifiers.
Granger causality test.
Performs pairwise Granger causality tests between (all possible) pairs of the group of series. If performed on series in a panel workfile, you may optionally choose to perform the Dumitrescu-Hurlin (2012) version of the test.
Syntax
group_name.cause(n, options)
Options
You must specify the number of lags n to use for the test by providing an integer in parentheses after the keyword. Note that the regressors of the test equation are a constant and the specified lags of the pair of series under test.
Panel Options
dh | Perform the Dumitrescu-Hurlin test. |
General Options:
prompt | Force the dialog to appear from within a program. |
p | Print output of the test. |
Examples
To compute Granger causality tests of whether GDP Granger causes M1 and whether M1 Granger causes GDP, you may enter the commands:
group g1 gdp m1
g1.cause(4)
The regressors of each test are a constant and four lags of GDP and M1.
The commands:
group macro m1 gdp r
macro.cause(12, p, dh)
print the result of six pairwise Dumitrescu-Hurlin causality tests for the three series in the MACRO group in a panel workfile.
Cross-references
See
“Granger Causality” for a discussion of Granger’s approach to testing hypotheses about causality. See
“Panel Causality Testing” for discussion of testing in panel settings.
Empirical distribution plot.
The
cdfplot command is no longer supported. See
distplot.Clear (i.e., replace with NAs) a contiguous block of observations in a group.
Syntax
group_name.clearcontents(start_point, col_range) n
where start_point specifies the first of n observations to clear. If n is negative, start_point specifies the last of |n| observations to clear. For dated workfiles, start_point should be entered as a date. For panels and undated workfiles, start_point should be an observation number.
The col_range option is used to specify the columns to be cleared in the group. It may take one of the following forms:
@all | Apply to all series in the group. |
col | Column number or letter (e.g., “2”, “B”). Apply to the series corresponding to the column. |
first_col[:]last_col | Colon-delimited range of columns (from low to high, e.g., “3:5”). Apply to all series corresponding to the column range. |
first_series[:]last_series | Colon-delimited range of columns (from low to high, e.g., “series01:series05”) specified by series names. Apply to all series corresponding to the column range. |
Examples
grp.clearcontents(1952Q2, 1) 10
clears 10 observations starting at 1952 quarter 2 in the first series in the group.
grp.clearcontents(10, gdp) -5
clears 5 observations ending at observation number 10 in the series GDP in the group.
grp.clearcontents(1990M2, @all) 8
clears 8 observations starting at February of 1990 in every series in the group.
Clear the contents of the history attribute for group objects.
Removes the group’s history attribute, as shown in the label view of the graph.
Syntax
group_name.clearhist
Examples
g1.clearhist
g1.label
The first line removes the history from the group G1, and the second line displays the label view of G1, including the now blank history field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Clear the contents of the remarks attribute.
Removes the group’s remarks attribute, as shown in the label view of the group.
Syntax
group_name.clearremarks
Examples
g1.clearremarks
g1.label
The first line removes the remarks from the group G1, and the second line displays the label view of G1, including the now blank remarks field.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Perform either (1) Johansen’s system cointegration test, (2) Engle-Granger or Phillips-Ouliaris single equation cointegration testing, or (3) Pedroni, Kao, or Fisher panel cointegration testing for the series in the group.
There are three forms for the coint command depending on which form of the test you wish to perform.
Johansen Cointegration Test Syntax
group_name.coint(options) [lag_spec] [@ x1 x2 x3 ...] [@exogsr sx1 sx2 sx3 ...] [@exoglr lx1 lx2 lx3 ...] [@exogboth bx1 bx2 bx3 ...]
uses the coint keyword followed by options, and optionally,
• a lag_spec consisting of one or more pairs of lag intervals, where the lag orders are for the differences in the error correction representation of the VEC, not the levels representation of the VAR.
• an “@”-sign or “@exogsr” followed by a list of exogenous variables in the short-run equation only
• “@exoglr” followed by a list of exogenous variables in the long-run relation only
• “@exogboth” followed by a list of exogenous variables in both the long-run relation and the short-run equations
(This type of cointegration testing may be used in a non-panel workfile except when performing Fisher combined testing using the Johansen framework.)
Note that the output for Johansen cointegration tests displays p-values for the rank test statistics. These p-values are computed using the response surface coefficients as estimated in MacKinnon, Haug, and Michelis (1999). The 0.05 critical values are also based on the response surface coefficients from MacKinnon-Haug-Michelis. Note: the reported critical values assume no exogenous variables other than an intercept and trend.
Options for the Johansen Test
Deterministic Trend Option
There are 8 different deterministic trend assumptions that you may specify using the “determ=arg” option.
These cases correspond to whether the intercept (“c”) and the trend (“t”) are either
• not included (“n”)
• in the long-run cointegrating relation only (“l”)
• in the short-run equation only (“s”)
• in both the long and short-run equations (“b”)
The values of arg are text shortcuts formed by joining a text shortcut for the intercept specification with a text shortcut for the trend specification.
The individual intercept and trend specifications are formed by joining the “c” and the “t” with the appropriate letter describing inclusion in the long and short-run equations.
For example,
• “cb” indicates that the constant is in both the long and short-run equation
• “tl” indicates that the trend is in the long-run cointegrating equation only
so that
• “cbtl” indicates that the constant is in both the long and short-run and the trend is in the long-run only
Using this convention (along with a special “none” option), we may easily describe options arguments for all 8 deterministic cases:
cntn, none | Case 1: No deterministic terms. Corresponding VAR model has no deterministic terms. |
cltn | Case 2: Restricted constant. Constant only in the cointegrating relations. Corresponding VAR has a constant. |
cbtn (default) | Case 3 (JHJ): Unrestricted constant Constant included both in the short-run equation and (artificially) in the cointegrating relations via orthogonalization. Corresponding VAR has a constant and trend. |
cstn | Case 3: Unrestricted constant Constant only in the short-run equation. Corresponding VAR has a trend. |
cbtl | Case 4 (JHJ): Unrestricted constant and restricted trend Constant included both in the short-run equation and (artificially) in the cointegrating relations via orthogonalization, and trend included only in the cointegrating relations. Corresponding VAR has a constant and trend. |
cstl | Case 4: Unrestricted constant and restricted trend Constant only in the short-run equation, and trend only in the cointegrating relation. Corresponding VAR has a trend. |
cbtb | Case 5 (JHJ): Unrestricted constant and trend Constant and trend both included in the short-run equation and (artificially) in the cointegrating relations via orthogonalization. Corresponding VAR has a constant, linear, and quadratic trend. |
csts | Case 5: Unrestricted constant and trend Constant and trend both included in the short-run equation. Corresponding VAR has a linear and quadratic trend. |
or you may use the “determsummary” option to compute tests under all deterministic assumptions.
Other Johansen Options
determsummary | Summarize all deterministic trend cases. |
restrict | Impose restrictions as specified by the “restspec=” option. |
restspec="spec" | Define the restricted VEC specification where spec is a space a space delimited list of VEC coefficient restrictions. |
m = integer, maxit = integer | Maximum number of iterations for restricted estimation (only valid if you choose the restrict option). |
c = scalar, cvg = scalar | Convergence criterion for restricted estimation. (only valid if you choose the restrict option). |
save = mat_name | Stores test statistics as a named matrix object. The save= option stores a 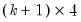 matrix, where 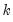 is the number of endogenous variables in the VAR. The first column contains the eigenvalues, the second column contains the maximum eigenvalue statistics, the third column contains the trace statistics, and the fourth column contains the log likelihood values. The i-th row of columns 2 and 3 are the test statistics for rank 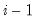 . The last row is filled with NAs, except the last column which contains the log likelihood value of the unrestricted (full rank) model. |
cvtype=ol | Display 0.05 and 0.01 critical values from Osterwald-Lenum (1992). This option reproduces the output from version 4. The default is to display critical values based on the response surface coefficients from MacKinnon-Haug-Michelis (1999). Note that the argument on the right side of the equals sign are letters, not numbers 0-1). |
cvsize=arg (default=0.05) | Specify the size of MacKinnon-Haug-Michelis (1999) critical values to be displayed. The size must be between 0.0001 and 0.9999; values outside this range will be reset to the default value of 0.05. This option is ignored if you set “cvtype=ol”. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Single Equation Test Syntax
group_name.coint(method=arg, options) [@determ determ_spec] [@regdeterm regdeterm_spec]
where
method=arg | Test method: Engle-Granger residual test (“eg”), Phillips-Ouliaris residual test (“po”). |
Cointegrating equation specifications that include a constant, linear, or quadratic trends, should use the “trend=” option to specify those terms. If any of those terms are in the stochastic regressors equations but not in the cointegrating equation, they should be specified using the “regtrend=” option.
Deterministic trend regressors that are not covered by the list above may be specified using the keywords @determ and @regdeterm. To specify deterministic trend regressors that enter into the regressor and cointegrating equations, you should add the keyword @determ followed by the list of trend regressors. To specify deterministic trends that enter in the regressor equations but not the cointegrating equation, you should include the keyword @regdeterm followed by the list of trend regressors.
Note that the p-values for the test statistics are based on simulations, and do not account for any user-specified deterministic regressors.
This type of cointegration testing may be used in a non-panel workfile. The remaining options for the single equation cointegration tests are outlined below.
Options for Single Equation Tests
Options for the Engle-Granger Test
The following options determine the specification of the Engle-Granger test (Augmented Dickey-Fuller) equation and the calculation of the variances used in the test statistic.
trend=arg (default=“const”) | Specification for the powers of trend to include in the cointegrating equation: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regtrend=arg (default=“none”) | Additional trends to include in the regressor equations (but not the cointegrating equation): None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Only trend orders higher than those specified by “trend=” will be considered. Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
lag=arg (default=“a”) | Method of selecting the lag length (number of first difference terms) to be included in the regression: “a” (automatic information criterion based selection), or integer (user-specified lag length). |
lagtype=arg (default=“sic”) | Information criterion or method to use when computing automatic lag length selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “msaic” (Modified Akaike), “msic” (Modified Schwarz), “mhqc” (Modified Hannan-Quinn), “tstat” (t-statistic). |
maxlag=integer | Maximum lag length to consider when performing automatic lag-length selection default= 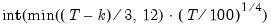 where 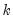 is the number of coefficients in the cointegrating equation. Applicable when “lag=a”. |
lagpval=number (default=0.10) | Probability threshold to use when performing automatic lag-length selection using a t-test criterion. Applicable when both “lag=a” and “lagtype=tstat”. |
nodf | Do not degree-of-freedom correct estimates of the variances. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Options for the Phillips-Ouliaris Test
The following options control the computation of the symmetric and one-sided long-run variances in the Phillips-Ouliaris test.
Basic Options
trend=arg (default=“const”) | Specification for the powers of trend to include in the cointegrating equation: None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
regtrend=arg (default=“none”) | Additional trends to include in the regressor equations (but not the cointegrating equation): None (“none”), Constant (“const”), Linear trend (“linear”), Quadratic trend (“quadratic”). Only trend orders higher than those specified by “trend=” will be considered. Note that the specification implies all trends up to the specified order so that choosing a quadratic trend instructs EViews to include a constant and a linear trend term along with the quadratic. |
nodf | Do not degree-of-freedom correct the coefficient covariance estimate. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
HAC Whitening Options
lag=arg (default=0) | Lag specification: integer (user-specified lag value), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum. |
HAC Kernel Options
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen). |
bw=arg (default=“nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
bwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Panel Test Syntax
group_name.coint(option)
The coint command tests for cointegration among the series in the group. This form of the command should be used with panel structured workfiles.
Options for the Panel Tests
For panel cointegration tests, you may specify the type using one of the following keywords:
Pedroni (default) | Pedroni (1994 and 2004). |
Kao | Kao (1999) |
Fisher | Fisher - pooled Johansen |
Depending on the type selected above, the following may be used to indicate deterministic trends:
const (default) | Include a constant in the test equation. Applicable to Pedroni and Kao tests. |
trend | Include a constant and a linear time trend in the test equation. Applicable to Pedroni tests. |
none | Do not include a constant or time trend. Applicable to Pedroni tests. |
determ=arg | Indicate deterministic trends as detailed above in
“Options for the Johansen Test”. Applicable to Fisher tests. |
Additional Options:
hac=arg (default=“bt”) | Method of estimating the frequency zero spectrum: “bt” (Bartlett kernel), “pr” (Parzen kernel), “qs” (Quadratic Spectral kernel). Applicable to Pedroni and Kao tests. |
bw=arg (default=“nw”) | Method of selecting the bandwidth, where arg may be “nw” (Newey-West automatic variable bandwidth selection), or a number indicating a user-specified common bandwidth. Applicable to Pedroni and Kao tests. |
lag=arg | For Pedroni and Kao tests, the method of selecting lag length (number of first difference terms) to be included in the residual regression. For Fisher tests, a pair of numbers indicating lag. |
infosel=arg (default=“sic”) | Information criterion to use when computing automatic lag length selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn). Applicable to Pedroni and Kao tests. |
maxlag=int | Maximum lag length to consider when performing automatic lag length selection, where int is an integer. The default is where 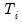 is the length of the cross-section. Applicable to Pedroni and Kao tests. |
disp=arg (default=500) | Maximum number of individual results to be displayed. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Examples
Johansen Test
gr1.coint(determsummary) 1 4
summarizes the results of the Johansen cointegration test for the series in the group GR1 for all specifications of trend. The test equation uses lags of up to order four.
Engle-Granger Test
gr1.coint(method=eg)
performs the default Engle-Granger test on the residuals from a cointegrating equation which includes a constant. The number of lags is determined using the SIC criterion and an observation-based maximum number of lags.
gr1.coint(method=eg, trend=linear, lag=a, lagtype=tstat, lagpval=.15, maxlag=10)
employs a cointegrating equation that includes a constant and linear trend, and uses a sequential t-test starting at lag 10 with threshold probability 0.15 to determine the number of lags.
gr1.coint(method=eg, lag=5)
conducts an Engle-Granger cointegration test on the residuals from a cointegrating equation with a constant, using a fixed lag of 5.
Phillips-Ouliaris Test
gr1.coint(method=po)
performs the default Phillips-Ouliaris test on the residuals from a cointegrating equation with a constant, using a Bartlett kernel and Newey-West fixed bandwidth.
gr1.coint(method=po, bw=andrews, kernel=quadspec, nodf)
estimates the long-run covariances using a Quadratic Spectral kernel, Andrews automatic bandwidth, and no degrees-of-freedom correction.
gr1.coint(method=po, trend=linear, lag=1, bw=4)
estimates a cointegrating equation with a constant and linear trend, and performs the Phillips-Ouliaris test on the residuals by computing the long-run covariances using AR(1) prewhitening, a fixed bandwidth of 4, and the Bartlett kernel.
Panel Tests
For a panel structured workfile,
grp1.coint(pedroni,maxlag=3,infosel=sic)
performs Pedroni’s residual-based panel cointegration test with automatic lag selection with a maximum lag limit of 3. Automatic selection based on Schwarz criterion.
Cross-references
See
“Cointegration Testing” for details on the various cointegration tests.
Creates a copy of the group.
Creates either a named or unnamed copy of the group.
Syntax
group_name.copy
group_name.copy dest_name
Examples
g1.copy
creates an unnamed copy of the group G1.
g1.copy g2
creates G2, a copy of the group G1.
Cross-references
Compute covariances, correlations and other measures of association for the series in a group.
You may compute measures related to Pearson product-moment (ordinary) covariances and correlations, Spearman rank covariances, or Kendall’s tau along with test statistics for evaluating whether the correlations are equal to zero.
Syntax
group_name.cor(options) [keywords [@partial z1 z2 z3...]]
You should specify keywords indicating the statistics you wish to display from the list below, optionally followed by the keyword @partial and a list of conditioning series or groups (for the group view), or the name of a conditioning matrix (for the matrix view). In the matrix view setting, the columns of the matrix should contain the conditioning information, and the number or rows should match the original matrix.
You may specify keywords from one of the four sets (Pearson correlation, Spearman correlation, Kendall’s tau, Uncentered Pearson) corresponding the computational method you wish to employ. (You may not select keywords from more than one set.)
If you do not specify
keywords, EViews will assume “corr” and compute the Pearson correlation matrix. Note that
Group::cor is equivalent to the
Group::cov command with a different default setting.
Pearson Correlation
cov | Product moment covariance. |
corr | Product moment correlation. |
sscp | Sums-of-squared cross-products. |
stat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
prob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Spearman Rank Correlation
rcov | Spearman’s rank covariance. |
rcorr | Spearman’s rank correlation. |
rsscp | Sums-of-squared cross-products. |
rstat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
rprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Kendall’s tau
taub | Kendall’s tau-b. |
taua | Kendall’s tau-a. |
taucd | Kendall’s concordances and discordances. |
taustat | Kendall’s score statistic for evaluating whether the Kendall’s tau-b measure is zero. |
tauprob | Probability under the null for the score statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Uncentered Pearson
ucov | Product moment covariance. |
ucorr | Product moment correlation. |
usscp | Sums-of-squared cross-products. |
ustat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
uprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Note that cases, obs, and wgts are available for each of the methods.
Options
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev” | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction to account for estimated means (for centered specifications), and any partial conditioning variables. |
multi=arg (default=“none”) | Adjustment to p-values for multiple comparisons: none (“none”), Bonferroni (“bonferroni”), Dunn-Sidak (“dunn”). |
outfmt=arg (default=“single”) | Output format: single table (“single”), multiple table (“mult”), list (“list”), spreadsheet (“sheet”). Note that “outfmt=sheet” is only applicable if you specify a single statistic keyword. |
out=name | Basename for saving output. All results will be saved in Sym matrices named using keys (“COV”, “CORR”, “SSCP”, “TAUA”, “TAUB”, “CONC” (Kendall’s concurrences), “DISC” (Kendall’s discordances), “CASES”, “OBS”, “WGTS”) appended to the basename (e.g., the covariance specified by “out=my” is saved in the Sym matrix “MYCOV”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
group grp1 height weight age
grp1.cor
displays a
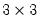
Pearson correlation matrix for the three series in
GRP1.
grp1.cor corr stat prob
displays a table containing the Pearson correlation, t-statistic for testing for zero correlation, and associated p-value, for the series in GRP1.
grp1.cor(pairwise) taub taustat tauprob
computes the Kendall’s tau-b, score statistic, and p-value for the score statistic, using samples with pairwise missing value exclusion.
grp1.cor(out=aa) cov @partial gender
computes the Pearson covariance for the series in GRP1 conditional on GENDER and saves the results in the symmetric matrix object AACOV.
Cross-references
See also
Group::cov. For simple forms of the calculation, see
@cor, and
@cov.
Display autocorrelation and partial correlations.
Displays the autocorrelation and partial correlation functions of the first series in the group, together with the Q-statistics and p-values associated with each lag.
Syntax
group_name.correl(n, options)
You must specify the largest lag n to use when computing the autocorrelations as the first option.
Options
d=integer (default=0) | Compute correlogram for specified difference of the data. |
prompt | Force the dialog to appear from within a program. |
p | Print the correlograms. |
Examples
gr1.correl(24)
Displays the correlograms of group GR1 for up to 24 lags.
Cross-references
See
“Autocorrelations (AC)” and
“Partial Autocorrelations (PAC)” for a discussion of autocorrelation and partial correlation functions, respectively.
Compute covariances, correlations and other measures of association for the series in a group.
You may compute measures related to Pearson product-moment (ordinary) covariances and correlations, Spearman rank covariances, or Kendall’s tau along with test statistics for evaluating whether the correlations are equal to zero.
Syntax
group_name.cov(options) [keywords [@partial z1 z2 z3...]]
You should specify keywords indicating the statistics you wish to display from the list below, optionally followed by the keyword @partial and a list of conditioning series or groups (for the group view), or the name of a conditioning matrix (for the matrix view).
You may specify keywords from one of the four sets (Pearson correlation, Spearman correlation, Kendall’s tau, Uncentered Pearson) corresponding the computational method you wish to employ. (You may not select keywords from more than one set.)
If you do not specify
keywords, EViews will assume “cov” and compute the Pearson covariance matrix. Note that
Group::cov is equivalent to the
Group::cor command with a different default setting.
Pearson Correlation
cov | Product moment covariance. |
corr | Product moment correlation. |
sscp | Sums-of-squared cross-products. |
stat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
prob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Spearman Rank Correlation
rcov | Spearman’s rank covariance. |
rcorr | Spearman’s rank correlation. |
rsscp | Sums-of-squared cross-products. |
rstat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
rprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Kendall’s tau
taub | Kendall’s tau-b. |
taua | Kendall’s tau-a. |
taucd | Kendall’s concordances and discordances. |
taustat | Kendall’s score statistic for evaluating whether the Kendall’s tau-b measure is zero. |
tauprob | Probability under the null for the score statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Uncentered Pearson
ucov | Product moment covariance. |
ucorr | Product moment correlation. |
usscp | Sums-of-squared cross-products. |
ustat | Test statistic (t-statistic) for evaluating whether the correlation is zero. |
uprob | Probability under the null for the test statistic. |
cases | Number of cases. |
obs | Number of observations. |
wgts | Sum of the weights. |
Note that cases, obs, and wgts are available for each of the methods.
Options
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev” | Weighting method (when weights are specified using “weight=”): frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations. Weights specified by “wgt=” are frequency weights for rank correlation and Kendall’s tau calculations. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
df | Compute covariances with a degree-of-freedom correction to account for estimated means (for centered specifications), and any partial conditioning variables. |
multi=arg (default=“none”) | Adjustment to p-values for multiple comparisons: none (“none”), Bonferroni (“bonferroni”), Dunn-Sidak (“dunn”). |
outfmt=arg (default= “single”) | Output format: single table (“single”), multiple table (“mult”), list (“list”), spreadsheet (“sheet”). Note that “outfmt=sheet” is only applicable if you specify a single statistic keyword. |
out=name | Basename for saving output. All results will be saved in Sym matrices named using keys (“COV”, “CORR”, “SSCP”, “TAUA”, “TAUB”, “CONC” (Kendall’s concurrences), “DISC” (Kendall’s discordances), “CASES”, “OBS”, “WGTS”) appended to the basename (e.g., the covariance specified by “out=my” is saved in the Sym matrix “MYCOV”). |
prompt | Force the dialog to appear from within a program. |
p | Print the result. |
Examples
group grp1 height weight age
grp1.cov
displays a
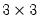
Pearson covariance matrix for the three series in
GRP1.
grp1.cov corr stat prob
displays a table containing the Pearson correlation, t-statistic for testing for zero correlation, and associated p-value, for the series in GRP1.
grp1.cov(pairwise) taub taustat tauprob
computes the Kendall’s tau-b, score statistic, and p-value for the score statistic, using samples with pairwise missing value exclusion.
grp1.cov(out=aa) cor @partial gender
computes the Pearson correlation for the series in GRP1 conditional on GENDER and saves the results in the symmetric matrix object AACORR.
Cross-references
See also
Group::cor. For simple forms of the calculation, see
@cor, and
@cov.
Displays cross correlations (correlograms) for a pair of series.
Syntax
group_name.cross(n,options)
You must specify the number of lags n to use in computing the cross correlations as the first option. Cross correlations will be computed for the first two series in the group.
Options
The following options may be specified inside the parentheses after the number of lags:
prompt | Force the dialog to appear from within a program. |
p | Print the cross correlogram. |
Examples
group grp1 log(m1) dlog(cpi)
grp1.cross(36)
displays the cross correlogram between the log of M1 and the first difference of the log of CPI, using up to 36 leads and lags.
equation eq1.arch sp500 c
eq1.makeresids(s) res_std
group g1 res_std^2 res_std
g1.cross(24)
The first line estimates a GARCH(1,1) model and the second line retrieves the standardized residuals. The third line creates a group and the fourth line plots the cross correlogram squared standardized residual and the standardized residual, up to 24 leads and lags. This correlogram provides a rough check of asymmetry in the ARCH effect.
Cross-references
Loads a dated data table template for the group.
Syntax
group_name.ddloadtmpl(options) template_name
template_name should be the name of a previously saved dated data table template.
Options
type=arg | Specify which settings to apply. type=trans loads the column group frequency, data display, table default transformation, and table default frequency conversion settings. type=appear loads the table default appearance settings. This includes the table default fonts, color, header options, label options, and formats. By default both types are loaded. |
series | Load series specific settings. This option is ignored if type=trans is used. |
Examples
group01.ddloadtmpl template1
loads all table settings from the template template1 and applies them to the dated data table of group GROUP01.
group01.ddloadtmpl(series) template1
loads both table settings and series specific settings from the template.
Cross-references
See
“Dated Data Table” for a description of dated data tables and formatting options.
See also
dtable and
ddtabopts.
Set row-specific options for dated date tables.
This proc sets row specific options for the group’s dated data table view. To set default settings for the dated data table, use the
ddtabopts proc.
Syntax
group_name.ddrowopts(series, row) args
You should provide integers to indicate the series and row number you wish to modify as an option to the command, followed by a list of arguments containing the display options for that row.
Arguments
transform(trans) | Set the transformation method for the row. trans can be: “l”(level), “d”(1 period diff), “yd”(year difference), “pc”(1 period % change), “pca”(1 period % chg-AR), “pcy”(year % chg), “tabdefault” (table default setting), “none” (don’t apply transformation) |
freqconv(conv) | Set the frequency conversion method for the row. conv can be “avgtran”(avg then transform), “tranavg”(transform then avg), “sumtran”(sum then transform), “first”(first period), “last”(last period), “tabdefault” (table default setting). |
format(fmt= new_format, units=new_units, prefix=” “, suffix=” “, +/-thousand, +/-comma, +/-parens) | Assign a custom prefix/suffix to the number, add a separator (comma or point) to denote thousands, replace a comma with a decimal point, or bracket negative numbers with parenthesis: fmt can be: “f[.prec]”(fixed decimal), “c[.prec]”(fixed characters), “auto”, “serformat”(series format). units can be: “N”(native), “P”(percent), “T”(thousands), “M”(millions), “B”(billions), “TR”(trillions) |
customrow(“string”) | Add a custom row header containing the quoted text string. To use a blank row, simply leave string empty. |
font( “name”, size, +/-b, +/-I, +/-s, +/-u) ) | Sets the font, size and style. name should be the name of the font, size should be an integer size value. You may use +b, +i, +s or +u to set bold, italic, strikeout or underline respectively. Use “tabdefault” to use the table default font setting. |
textcolor(arg) | Sets the color of the text. arg may be one of the predefined color keywords, or it may be specified using individual red-green-blue (RGB) components using the “@RGB” or “@HEX” functions. The arguments to the @RGB function are a set of three integers from 0 to 255, representing the RGB values of the color. The arguments to the “@HEX” function are a set of six characters representing the RGB values of the color in hexadecimal. Each two character set represents a red, green or blue component in the range '00' to 'FF'. For a description of the available color keywords see
“Color definitions”. |
rowlabel(label) | Sets a custom row label in place of the series name. |
Color definitions
color_arg specifies the color to be employed in the arguments above. The color may be specified using predefined color names, by specifying the individual red-green-blue (RGB) components using the special “@RGB” function, or by specifying the individual red-green-blue (RGB) components in hexadecimal using the special “@HEX” function.
The predefined colors are given by the keywords (with their RGB or HEX equivalents):
blue | @rgb(0, 0, 255) | @hex(0000ff) |
red | @rgb(255, 0, 0) | @hex(ff0000) |
ltred | @rgb(255, 168, 168) | @hex(ffa8a8) |
green | @rgb(0, 128, 0) | @hex(008000) |
black | @rgb(0, 0, 0) | @hex(000000) |
white | @rgb(255, 255, 255) | @hex(ffffff) |
purple | @rgb(128, 0, 128) | @hex(800080) |
orange | @rgb(255, 128, 0) | @hex(ff8000) |
yellow | @rgb(255, 255, 0) | @hex(ffff00) |
gray | @rgb(128, 128, 128) | @hex(808080) |
ltgray | @rgb(192, 192, 192) | @hex(c0c0c0) |
Examples
The following examples show the use of dtable, ddtabopts and ddrowopts together to customize dated display tables.
group cgrp cenergy cfood chealth
cgrp.dtable
cgrp.ddtabopts firstfreq(a) secfreq(none) display(year,1)
cgrp.ddrowopts(1,1) transform(l) format(fmt=f.1) customrow("Consumption Indicators")
cgrp.ddrowopts(1,2) transform(pc) format(fmt=f.2, parens) fillcolor(red)
cgrp.ddrowopts(2,2) transform(pcy) format(fmt=f.2) fillcolor(blue)
creates the group CGGRP from the series CENERGY, CFOOD AND CHEALTH, and displays the dated data table for that group. ddtapopts is used to set the first table frequency to annual and the second frequency to none, displaying one year of data per row.
The three ddrowopts commands set display options for CENERGY and CFOOD. For CENERGY the first row is transformed to levels, numbers are displayed to one decimal place, and row is placed above the series with the custom string "Consumption Indicators". The next command adds a red row to CENERGY with the data transformed to 1-period percent changes, rounded to two decimal places, surrounded by parentheses if negative. The last command adds a blue filled row to CFOOD containing 1-year percent changes rounded to two decimal places.
group fgrp houliab hounetworth houassets
fgrp.dtable
fgrp.ddtabopts firstfreq(q) secfreq(a)
fgrp.ddrowopts(1,1) freqconv(avgtran) format(units=t)
fgrp.ddrowopts(3,1) format(fmt=f.0) transform(d) textcolor(blue)
The ddtabopts command sets the table defaults to show blocks of quarterly and annual data in the dated data table. The ddrowopts commands change the way HOULIAB and HOUSALES are displayed in the dated data table view. HOULIAB is set to be averaged then transformed with the units set to thousands. HOUASSETS are set to zero decimal places, transformed to the first period difference and changed to a blue font color.
group ggpr govinv govpurchases govsav
ggpr.dtable
ggpr.ddtabopts qtrformat(qr) nalabel("NA") rowheader(+b)
ggpr.ddrowopts(1,1) transform(pca) freqconv(tabdefault)
ggpr.ddrowopts(2,1) transform(pc) customrow(" ")
ggpr.ddrowopts(3,2) transform(pcy)
The ddtabopts command sets the table defaults to show the quarter in short case roman numerals, then adds an "NA" to any missing data, it also bold the row headers. The ddrowopts command transforms the GOVINV display to percent change annual rate and sets the frequency conversion method to the table default. The proc also sets GOVPURCHASES to percent change, adds a blank row above the data, and adds a transformed 1-year percent change row to GOVSAV.
group igrp natincome persincome dispincome
igrp.dtable
igrp.ddtabopts font("arial",10) colheader(b)
igrp.ddrowopts(1,1) transform(pca) format(fmt=f.1) textcolor(red)
igrp.ddrowopts(2,1) transform(pca) format(fmt=parens)
igrp.ddrowopts(3,1) transform(pca) customstring("Disp. Income")
The ddtabopts command sets the font to Arial size 10 and bolds the column header. For the series NATINCOME the ddrowopts command transforms the series to percent change annual rate, sets the numerical format to one decimal place, and sets the text color to red. For PERSINCOME the command adds a parenthesis for negative numbers, and for DISPINCOME it adds a custom row above the series containing the text heading “Disp. Income”.
Cross-references
See
“Dated Data Table” for a description of dated data tables and formatting options.
See also
dtable and
ddtabopts.
Saves the current dated data table settings as a new template.
Syntax
group_name.ddsavetmpl(options) template_name
Options
overwrite | Overwrite an existing template with the same name. Without this option naming conflicts will result in an error. |
Examples
group01.ddsavetmpl template1
saves the current dated data template settings of group GROUP01 as the new template template1.
Cross-references
See
“Dated Data Table” for a description of dated data tables and formatting options.
See also
dtable and
ddtabopts.
Set table default options for dated data tables.
Specifies the table default options for the group’s dated data table view. To set row specific options that override the defaults, use the
ddrowopts proc.
Syntax
group_name.ddtabopts args
Arguments
display(arg, n) | Specify the data to display in each table row. arg can be “first”, “last” or “year”. “first” or “last” will display annual totals, plus the first, or last, n observations in each row. “year” will display observations for n years of data per row. |
firstfreq(freq) | Sets the frequency for the first column grouping: freq can be “n”(native), “a”(annual), “q”(quarterly), “m”(monthly). |
secfreq(freq) | Sets the frequency for the second column grouping: freq can be “none” (none), “n”(native), “a”(annual), “q”(quarterly), “m”(monthly). |
nalabel(“arg”) | Sets the label for NA values to arg. |
+/-displayname | Use display names as default labels. |
transform(row, trans) | Set the transformation method for row row. trans can be: “l”(level), “d”(1 period diff), “yd”(year difference), “pc”(1 period % change), “pca”(1 period % chg-AR), “pcy”(year % chg). |
freqconv(row, conv) | Set the frequency conversion method for the specified row. conv can be “avgtran” (avg then transform), “tranavg” (transform then avg), “sumtran” (sum then transform), “first” (first period), “last”(last period). |
format(fmt= new_format, units=new_units, prefix=” “, suffix=” “, +/-thousand, +/-comma, +/-parens) | Assign a custom prefix/suffix to the number, add a separator (comma or point) to denote thousands, replace a comma with a decimal point, or bracket negative numbers with parenthesis: fmt can be: “f[.prec]”(fixed decimal), “c[.prec]”(fixed characters), “auto”, “serformat”(series format). units can be: “N”(native), “P”(percent), “T”(thousands), “M”(millions), “B”(billions), “TR”(trillions) |
colheader(+/-b, +/- i) | Sets column headers to bold or italic style. |
rowheader(+/-b, +/- i) | Sets row headers to bold or italic style. |
fillcolor(arg) | Set the table row background color to that specified by arg. arg may be one of the predefined color keywords, or it may be specified using individual red-green-blue (RGB) components using the “@RGB” or “@HEX” functions. The arguments to the @RGB function are a set of three integers from 0 to 255, representing the RGB values of the color. The arguments to the “@HEX” function are a set of six characters representing the RGB values of the color in hexadecimal. Each two character set represents a red, green or blue component in the range '00' to 'FF'. For a description of the available color keywords see
“Color definitions”. |
altfillcolor(colorspec) | Set the table alternate row background color to colorspec. colorspec may consist of an @rgb(r,g,b) or @hex specification or it may be the name of a basic color such as “white”, “blue”, “red”, “black”, etc. |
font(row, “name”, size, +/–b, +/–I, +/–s, +/–u) | Sets the font, size and style. name should be the quoted name of the font, size should be an integer size value. You may use +b, +i, +s or +u to set bold, italic, strikeout or underline respectively. |
yrformat(arg) | Sets the date format for year date labels. arg may be “YYYY” (4-digit years) or “YY” (2 digit years). |
qtrformat(arg) | Sets the date format for quarterly date labels. arg may be “QR” (upper-case Roman numerals), “qr” (lower-case Roman numerals), “[Q]Q” (“Q” followed by the quarter number), “Q” (quarter number), “Mon” (3 letter month abbreviation for first month in quarter), “Month” (full month name for first month in quarter). |
monformat(arg) | Sets the date format for monthly date labels. arg may be “[M]mm” (“M” followed by month number), “mm” (month number), “MM” (month number with preceding zero), “[M]MM” (“M” followed by month number with preceding zero) “Mon” (3 letter month abbreviation), “Month” (full month name), “M” (upper-case first letter of month name), or “m” (lower-case first letter of month name). |
qtryrformat(arg) | Sets the joint date format for quarter and year. Only applicable if “Display(first)” or “Display(last)” is used. arg may be: “YYYY[q]Q]”, “YYYY[Q]Q”, “YYYY:Q”, “YY[q]Q”, “YY[Q]Q”, “YY:Q”, “YYYY QR”, “YYYYqr”, “YYYY qr”, “YY QR”, “YYqr”, “YY qr”, “Mon YYYY”, “Mon YY”, or “Month YYYY”. See description of “yrformat” and “qtrformat” above for details. |
monyrformat(arg) | Sets the joint date format for month and year. Only applicable if “Display(first)” or “Display(last)” is used. arg may be: “YYYY[m]mm]”, “YYYY[M]mm”, “YYYY[m]MM”, “YYYY:M”, “YY[m]mm”, “YY[M]mm”, “YY[m]MM”, “YY:mm”, “YY:MM”, “Mon YYYY”, “Mon YY”, “Month YYYY”, “Month YY”, “YYMon”, or “YY-Mon”. See description of “yrformat” and “monformat” above for details on each. |
+/– endperiod | Use end of period date labels. |
Color definitions
color_arg specifies the color to be employed in the arguments above. The color may be specified using predefined color names, by specifying the individual red-green-blue (RGB) components using the special “@RGB” function, or by specifying the individual red-green-blue (RGB) components in hexadecimal using the special “@HEX” function.
The predefined colors are given by the keywords (with their RGB and HEX equivalents):
blue | @rgb(0, 0, 255) | @hex(0000ff) |
red | @rgb(255, 0, 0) | @hex(ff0000) |
ltred | @rgb(255, 168, 168) | @hex(ffa8a8) |
green | @rgb(0, 128, 0) | @hex(008000) |
black | @rgb(0, 0, 0) | @hex(000000) |
white | @rgb(255, 255, 255) | @hex(ffffff) |
purple | @rgb(128, 0, 128) | @hex(800080) |
orange | @rgb(255, 128, 0) | @hex(ff8000) |
yellow | @rgb(255, 255, 0) | @hex(ffff00) |
gray | @rgb(128, 128, 128) | @hex(808080) |
ltgray | @rgb(192, 192, 192) | @hex(c0c0c0) |
Examples
group cgrp cenergy cfood chealth
cgrp.dtable
cgrp.ddtabopts firstfreq(a) secfreq(none) display(year,1)
creates the group CGRP from the series CENERGY, CFOOD AND CHEALTH, and then it displays the dated data table for that group. ddtapopts is used to set the first table block frequency to annual and the second frequency to none, with one year of data displayed in each row.
group fgrp houliab hounetworth houassets
fgrp.dtable
fgrp.ddtabopts +displayname firstfreq(q) secfreq(a) colheader(i) font("Calibri",10) altfillcolor(yellow) qtrformat([Q]Q)
The ddtabopts command sets the table options for the group FGRP to show the display-names of each series in place of the series names, sets the first block frequency to quarterly and the second to annual, sets the column header style to italics, changes the font to Calibri size 10, sets the alternative row color to yellow, and sets the display for quarterly data to “Q[q]”.
group govgrp govinv govpurchases govsav
govgrp.dtable
govgrp.ddtabopts qtrformat(qr) nalabel("NA") rowheader(+b)
creates the group GOVGRP out of the series GOVINV, GOVPURCHASES, and GOVSAV and then the dated data table. The ddtabopts command is set to show the quarter in short case roman numerals, then adds an "NA" to any missing data, it also bold the row headers.
group hgrp starts singlestarts multistarts
hgrp.dtable
hgrp.ddtabopts firstfreq(a) secfreq(none) +displayname fillcolor(@rgb(205,201,201)) yrformat(YY) format(units=n, fmt=f.2)
The ddtabopts command sets table default options for the group HGRP, with the first column grouping frequency as annual and the second grouping to none. The table defaults will show displaynames in place of series names, will use a light gray row fill color specified by RGB. The year format is set to show only the last two digits of the year and the numerical display format is set to native with two decimal places.
group incgrp natincome persincome dispincome
incgrp.dtable
incgrp.ddtabopts font("arial",10) colheader(b)
sets the table default font to size 10 Arial and specifies bold column headers.
Cross-references
See
“Dated Data Table” for a description of dated data tables and formatting options.
See also
dtable and
ddrowopts.
Delete observations from a group.
Syntax
group_name.deleteobs(col, row) [num_obs]
where col is an integer specifying the column to delete, row is an integer specifying the first row to delete, and num_obs specifies the number of observations to delete from the table.
Examples
group1.deleteobs(2, 5) 15
deletes fifteen observations from the second series in the group starting at the fifth row.
Display table, graph, or spool output in the group object window.
Display the contents of a table, graph, or spool in the window of the group object.
Syntax
group_name.display object_name
Examples
group1.display tab1
Display the contents of the table TAB1 in the window of the object GROUP1.
Cross-references
Most often used in constructing an EViews Add-in. See
“Custom Object Output” .
Display name for the group object.
Attaches a display name to a group object which may be used to label output in tables and graphs in place of the standard group object name.
Syntax
group_name.displayname display_name
Display names are case-sensitive, and may contain a variety of characters, such as spaces, that are not allowed in group object names.
Examples
grp1.displayname Hours Worked
grp1.label
The first line attaches a display name “Hours Worked” to the group object GRP1, and the second line displays the label view of GRP1, including its display name.
Cross-references
See
“Labeling Objects” for a discussion of labels and display names.
Save a matrix containing distribution plot data computed from the group.
Saves the data used to display the kernel regression, nearest neighbor regression, or empirical quantile-quantile plot to the workfile.
Syntax
group_name.distdata(dtype=dist_type, dist_options) matrix_name_pattern
saves the distribution plot data specified by dist_type where dist_type must be one of the following keywords:
kernfit | Kernel regression (default). |
nnfit | Nearest neighbor (local) regression. |
empqq | Empirical quantile-quantile plot. |
The matrix_name_pattern is used to define a naming pattern for the output matrices; if the pattern is “NAME”, the resulting matrices will be named “NAME01”, “NAME02”, … and so on, using the next available name.
Options
For the first two types (“kernfit” and “nnfit”),
dist_options are any of the distribution type-specific options described in
“Kernfit Options” and
“Nnfit Options”, respectively. The empirical quantile-quantile plot type (“empqq”) takes the options described in
qqplot under
“Empirical Options”.
Note that the graph display specific options such as “fill,” “nofill,” “leg,” and “noline” are not relevant for this procedure.
In addition, you may use the “mult” option to specify multiple series handling
mult = mat_type | Multiple series or column handling: where mat_type may be: “pairs” or “p” - pairs, “mat” or “m” - scatterplot matrix, “lower” or “l” - lower triangular matrix. |
and the “prompt” option to force the dialog display
prompt | Force the dialog to appear from within a program. |
Examples
group g w x y z
g.distdata(mult=first, dtype=kernel, k=e, ngrid=100) m
creates a group called G from the series X, Y and Z, then creates three matrices, M01, M02 and M03, where the first matrix contains the kernel fit (with an Epanechnikov kernel and 100 grid points) of W on X, the second contains the fit of W on Y, and the third matrix contains the kernel fit of Won Z.
g.distdata(mult=pairs, dtype=local, b=0.3, d=1, neval=100, s) n
creates two matrices, N1 and N2, where N1 contains the nearest neighbor fit of W on X computed using a bandwidth of 0.3 and polynomial degree of 1, 100 evaluation points and symmetric neighbors, and N2 contains the data for the nearest neighbor fit of Y on Z.
group g.drop z
g.distdata(mult=all, dtype=empqq, q=r) mat
drops Z from the group, then creates 3 matrices; MAT01, MAT02, MAT03, where MAT01 contains the empirical quantile-quantile for W and X, computed using the rankit quantile method, and MAT02 contains the qq-plot data for W and Y, and MAT03 contains the qq-plot data for X and Y.
Cross-references
For a description of distribution graphs and quantile-quantile graphs, see
“Analytical Graph Types”.
Drops series from a group.
Syntax
group_name.drop ser1 [ser2 ser3 ...]
List the series to be dropped from the group object.
Examples
group gdplags gdp(-1 to -4)
gdplags.drop gdp(-4) gdp(-3)
drops the two series GDP(-4) and GDP(-3) from the group GDPLAGS.
Cross-references
See
“Groups” for additional discussion of groups.
Dated data report table.
This group view is designed to make tables for reporting and presenting data, forecasts, and simulation results. You can display various transformations and various frequencies of the data in the same table.
The dtable view is currently available only for annual, semi-annual, quarterly, or monthly workfiles.
Syntax
group_name.dtable(options)
Options
Examples
freeze(report) group1.dtable
freezes the dated table view of GROUP1 and saves it as a table object named REPORT.
Cross-references
See
“Dated Data Table” for a description of dated data tables and formatting options.
Duplicate observations display for observations in the group.
Syntax
group_name.dups(opts)
A duplicate is defined as two observations with the same values for all series in the group.
By default, EViews displays a summary table showing the number of duplicate groups of a given size, but you may use the options to display an alternative view.
Of particular note is that the spreadsheet and individual duplicates displays are interactive - clicking on rows in one will open the display to show the other. Thus, clicking on a duplicate in the spreadsheet view will jump to show all of the observations that share that duplicate. Similarly, clicking on an observation in the shared individual duplicates view will jump to the corresponding observation in the full spreadsheet.
Options
graph | Display observation graph showing duplicates. |
sheet | Display spreadsheet view of duplicates. |
individ | Display first individual duplicates. |
Examples
grp1.dups
displays the duplicates summary for the group GRP1.
grp1.dups(sheet)
displays a spreadsheet showing highlighted duplicates.
Cross-references
Compute frequency tables for the series in a group.
The freq command performs a one-way or N-way frequency tabulation.
• When used with a group containing a single series, freq performs a one-way frequency tabulation.
• When used with a group containing multiple series, freq produces an N-way frequency tabulation for all of the series in the group.
Frequencies are computed for the current sample of observations. Observations with NAs are excluded unless included by option. You may use options to control automatic binning (grouping) of numeric values and the order of the entries of the table.
Syntax
group_name.freq(options)
Options
Options common to both one-way and N-way frequency tables
dropna (default) / keepna | [Drop/Keep] NA as a category. |
v=integer (default=1000) | Make bins if the number of distinct values or categories exceeds the specified number. |
nov | Do not make bins on the basis of number of distinct values; ignored if you set “v=integer.” |
a=number | (optional) Make bins if average count per distinct value is less than the specified number. |
b=integer (default=50) | Maximum number of categories to bin into if performing automatic binning. |
n, obs, count (default) | Display frequency counts. |
nocount | Do not display frequency counts. |
nolimit | Remove prompt warning for continuing when the total number of cells is very large. |
sort=arg (default=“lohi”) | Sort order for entries in the frequency table: high data value to low ("hilo"), low data value to high ("lohi" –default), high frequency to low ("freqhilo"), low frequency to high ("freqlohi"). |
prompt | Force the dialog to appear from within a program. |
p | Print the table. |
Options for one-way tables
total (default) / nototal | [Display / Do not display] totals. |
pct (default) / nopct | [Display / Do not display] percent frequencies. |
cum (default) / nocum | (Display/Do not) display cumulative frequency counts/percentages. |
Options for N-way tables
table (default) | Display in table mode. |
list | Display in list mode. |
rowm (default) / norowm | [Display / Do not display] row marginals. |
colm (default) / nocolm | [Display / Do not display] column marginals. |
tabm (default) / notabm | [Display / Do not display] table marginals—only for more than two series. |
subm (default) / nosubm | [Display / Do not display] sub marginals—only for “l” option with more than two series. |
full (default) / sparse | (Full/Sparse) tabulation in list display. |
totpct / nototpct (default) | [Display / Do not display] percentages of total observations. |
tabpct / notabpct (default) | [Display / Do not display] percentages of table observations—only for more than two series. |
rowpct / norowpct (default) | [Display / Do not display] percentages of row total. |
colpct / nocolpct (default) | [Display / Do not display] percentages of column total. |
exp / noexp (default) | [Display / Do not display] expected counts under full independence. |
tabexp / notabexp (default) | [Display / Do not display] expected counts under table independence—only for more than two series. |
test (default) / notest | [Display / Do not display] tests of independence. |
Examples
group g1 hrs
g1.freq(nov, noa)
tabulates each value (no binning) of HRS in ascending value order with counts, percentages, and cumulatives.
group g2 inc
g2.freq(v=200, b=100, keepna, noa)
tabulates INC including NAs. The values will be binned if INC has more than 200 distinct values; EViews will create at most 100 equal value-width bins. The number of bins may be less than 100.
group labor lwage gender race
labor.freq(v=10, norowm, nocolm)
displays tables of binned LWAGE against GENDER for each bin/value of RACE. The table will not contain row and column marginals.
labor.freq(v=10, norowm, nocolm, sort=freqhilo)
displays the same table with the table rows and columns ordered by values with highest frequency to lowest.
Cross-references
See
“One-Way Tabulation” and
“N-Way Tabulation” for a discussion of frequency tables.
Declare a group object containing a group of series.
Syntax
group group_name ser1 ser2 [ser3 ...]
Follow the group name with a list of series to be included in the group.
The wildcard operator, *, may be used as part of the series list to include many series at once. The keywords AND or NOT can be used to specify certain series should not be included in the group.
Examples
group g1 gdp cpi inv
group g1 tb3 m1 gov
g1.add gdp cpi
The first line creates a group named
G1 that contains three series
GDP,
CPI, and
INV. The second line redeclares group
G1 to contain the three series
TB3,
M1, and
GOV. The third line adds two series
GDP and
CPI to group
G1 to make a total of five series. See
Group::add.
group rhs d1 d2 d3 d4 gdp(0 to -4)
ls cons rhs
ls cons c rhs(6)
The first line creates a group named RHS that contains nine series. The second line runs a linear regression of CONS on the nine series in RHS. The third line runs a linear regression of CONS on C and only the sixth series GDP(-1) of RHS.
group g2 us_*
This line creates a group named G2 that contains any series whose name starts with the characters US_.
group g3 * not resid
This command makes a group, G3, containing all series in the workfile except for the resid series.
group g4 a* and *1
Makes a group named G4 containing all series whose names begin with the letter A and end with L.
group g5 a* b* not *1 *2
This line makes a group, G5, containing all series whose names begin with either letter A or B and do not end with either 1 or 2.
group g6 g1 and g2
Makes a group named G6 containing all series that are both in group G1 and group G2 (i.e. the intersection of the two groups).
Cross-references
See
“Groups” for additional discussion.
Shift the observations of the series in the group up or downwards, inserting blank observations.
Syntax
group_name.insertobs(startpoint, col_range) n
Where startpoint specifies the first or last observation from which the observations are shifted. For dated workfiles, startpoint should be entered as a date. For panels and non-dated workfiles startpoint should be an observation number.
The col_range option is used to describe the columns to be shifted in the group. It may take one of the following forms:
@all | Apply to all series in the group. |
col | Column number or letter (e.g., “2”, “B”). Apply to the series corresponding to the column. |
first_col[:]last_col | Colon delimited range of columns (from low to high, e.g., “3:5”). Apply to all series corresponding to the column range. |
first_series[:]last_series | Colon delimited range of columns (from low to high, e.g., “series01:series05”) specified by the series names. Apply to all series corresponding to the column range. |
n specifies the number of observations shifted.
Examples
g.insertobs(1952q2, 1) 2
Inserts 2 new observations beginning at observation 1952 quarter 2 into the first series in the group. The previous value associated with 1952Q2 for that series will now correspond to 1952Q4.
g.insertobs(10, gdp) -5
Inserts 5 new observations to the series GDP ending at observation number 10.
g.insertobs(1990m2, @all) 8
Inserts 8 new observations beginning at February 1990 for all series in the group.
Scatterplot with bivariate kernel regression fit.
The
kerfit command is no longer supported. See
scat.
Display or change the label view of a group, including the last modified date and display name (if any).
As a procedure, label changes the fields in the group label.
Syntax
group_name.label
group_name.label(options) [text]
Options
The first version of the command displays the label view of the group. The second version may be used to modify the label. Specify one of the following options along with optional text. If there is no text provided, the specified field will be cleared.
c | Clears all text fields in the label. |
d | Sets the description field to text. |
s | Sets the source field to text. |
u | Sets the units field to text. |
r | Appends text to the remarks field as an additional line. |
p | Print the label view. |
Examples
The following lines replace the remarks field of G1 with “Data from CPS 1988 March File”:
g1.label(r)
g1.label(r) Data from CPS 1988 March File
To append additional remarks to G1, and then to print the label view:
g1.label(r) Log of hourly wage
g1.label(p)
To clear and then set the units field, use:
g1.label(u) Millions of bushels
Cross-references
See
“Labeling Objects” for a discussion of labels.
Scatterplot with bivariate fit.
The
linefit command is no longer supported. See
scat.
Compute the symmetric, one-sided, or strict one-sided long-run covariance matrix for a group of series.
Syntax
Group View: group_name.lrcov(options)
Options
window=arg | Type of long-run covariance to compute: “sym” (symmetric), “lower” (lower - lags in columns), “slower” (strict lower - lags only), “upper” (upper - leads in columns), “supper” (strict upper - leads only) |
noc | Do not remove means (center data). |
rwgt=arg | Row weights. |
out=arg | Name of output sym or matrix (optional). |
panout=arg | Name of panel output matrix (optional). |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Whitening Options
lag=arg (default=0) | Lag specification: integer (user-specified number of lags), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 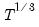 . |
Kernel Options
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett, default), “bohman” (Bohman), “daniell” (Daniell), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen), “user” (User-specified; see “kernwgt=” below). |
kernwgt=vector | User-specified kernel weight vector (if “kern=user”). |
bw=arg (default=”nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
bwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
Examples
grp1.lrcov(out=outsym)
computes the symmetric long-run covariance of the series in the group GRP1 and saves the results in the output sym matrix OUTSYM.
xgrp.lrcov(kern=quadspec, bw=andrews, rwgt=res)
computes the long-run covariance of the series in the group XGRP using the quadratic spectral kernel, Andrews automatic bandwidth, and the row-weight series RES.
xgrp.lrcov(kern=quadspec, lag=1, bw=andrews, rwgt=res)
performs the same calculation but uses VAR(1) prewhitening prior to computing the kernel estimator.
xgrp.lrcov(kern=none, window=upper, lag=a, infosel=aic, bw=andrews, rwgt=res)
computes parametric VAR estimates of the upper long-run covariance using an AIC based automatic bandwidth selection method.
Cross-references
Push updates to OLE linked objects in open applications.
Syntax
group_name.olepush
Cross-references
See
“Object Linking and Embedding (OLE)” for a discussion of using OLE with EViews.
Save the scores from a principal components analysis of the series in a group.
Syntax
group_name.makepcomp(options) output_list
where the
output_list is a list of names identifying the saved components. EViews will save the first
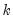
components corresponding to the
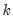
elements in
output_list, up to the total number of series in the group.
Options
scale=arg (default=“normload”) | Diagonal matrix scaling of the loadings and the scores: normalize loadings (“normload”), normalize scores (“normscores”), symmetric weighting (“symmetric”), user-specified (arg=number). |
cpnorm | Compute the normalization for the score so that cross-products match the target (by default, EViews chooses a normalization scale so that the moments of the scores match the target). |
eigval=vec_name | Specify name of vector to hold the saved the eigenvalues in workfile. |
eigvec=mat_name | Specify name of matrix to hold the save the eigenvectors in workfile. |
prompt | Force the dialog to appear from within a program. |
Covariance Options
cov=arg (default=“corr”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), uncentered ordinary correlation (“ucorr”). Note that Kendall’s tau measures are not valid methods. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev” | Weighting method: frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations where “weights=” is specified. Weights for rank correlation and Kendall’s tau calculations are always frequency weights. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
partial=arg | Compute partial covariances conditioning on the list of series specified in arg. |
df | Compute covariances with a degree-of-freedom correction accounting for the mean (for centered specifications) and any partial conditioning variables. The default behavior in these cases is to perform no adjustment ( e.g. – compute sample covariance dividing by 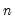 rather than 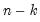 ). |
Examples
grp1.makepcomp comp1 comp2 comp3
saves the first three principal components (in normalized loadings form) to the workfile. The components will have variances that are proportional to the eigenvalues.
grp1.makepcomp(scale=normscore) comp1 comp2 comp3
normalizes the scores so that the resulting series have variances that are equal to 1.
You may change the scaling for the normalized components so that the cross-products equal 1, using the cpnorm option:
grp1.makepcomp(scale=normscore, cpnorm) comp1 comp2 comp3
Cross-references
See
“Saving Component Scores” for further discussion.
See
Group::pcomp for tools to display the principal components results for the series in the group.
Create system from a group.
Syntax
group_name.makesystem(options) [x1 x2 x3 ...] [@eqreg w1 w2 ...] [@inst z1 z2 ...] [@eqinst z3 z4 ...]
Creates a system of equations out of the variables in the group. Each series in the group will be used as the dependent variable in an equation. The [x1 x2 x3 ...] list consists of regressors with common coefficients in the system. The @eqreg list consists of regressors with different coefficients in each equation. The list of variables that follow @inst are the common instruments. The list of variables that follow @eqinst are the equation specific instruments.
Options
name=name | Specify name for the system object. |
ytrans=arg | Dependent variable transformation: none (default), log (“log”), difference (“d”), difference of logs (“dlog”), one percentage change in decimal (“pch”), one-period percentage change—annualized, in percent (“pcha”), one-year percentage change in decimal (“pchy”). |
prompt | Force the dialog to appear from within a program. |
Examples
grp1.makesystem(name=sys1) c x1 x2 @inst z1 z2 z3
creates a system named SYS1 with the series in GRP1 as the dependent variables and a common intercept and coefficients on X1 and X2, with common instruments Z1, Z2, and Z3.
grp1.makesystem(name=sys2) x1 @eqreg c x2 @inst z1 z2 @eqinst z3
creates a system named SYS2 with a common coefficient for X1 and a different intercept and coefficient for X2 for each equation. There are common intercepts Z1 and Z2, and an equation specific instrument Z3.
Cross-references
See
“System Estimation” for a discussion of system objects in EViews.
Whiten the series in the group.
Estimate a VAR(
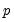
) for the series in the group, compute the residuals, and save the results into whitened series.
Syntax
Group View: group_name.makewhiten(options) out_specification
where out_specification is either a list of names for the output series, one per series in the original group, or is a wildcard expression. Note that wildcards may not be used if the original group contains series expressions.
Options
grp=arg | Name of group to hold output series (optional). |
lag=arg (default=1) | Lag specification: integer (user-specified number of lags), “a” (automatic selection). |
noc | Do not remove means (center data) prior to whitening. |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn). |
maxlag=integer | Maximum lag-length for automatic selection (optional). The default is an observation-based maximum of the integer portion of 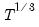 . |
prompt | Force the dialog to appear from within a program. |
Examples
grp1.makewhiten(grp=wht, lag=a, infosel=sic, maxlag=10) *a
whitens the series in GRP1 using a VAR with auto-selected number of lags based on the SIC information criterion and a maximum of 10 lags. The resulting series are named using the wildcard expression “*a” in the named group WHT.
grp2.makewhiten(noc, lag=5) *a
whitens the series in GRP2 using a no-constant VAR and 5 lags.
Cross-references
See
“Make Whitened” for detail.
Display the members of the group.
Syntax
Group View: group_name.members
Examples
grp1.members
Cross-references
See
“Group Members” for additional detail.
Scatterplot with bivariate nearest neighbor fit.
The
nnfit command is no longer supported. See
scat.
Principal components analysis.
Syntax
group_name.pcomp(options) [indices]
where the elements to display in loadings, scores, and biplot graph form (“out=loadings”, “out=scores” or “out=biplot”) are given by the optional indices, (e.g., “1 2 3” or “2 3”). If indices is not provided, the first two elements will be displayed.
Basic Options
out=arg (default=“table”) | Output type: eigenvector/eigenvalue table (“table”), eigenvalues graph (“graph”), loadings graph (“loadings”), scores graph (“scores”), biplot (“biplot”). |
eigval=vec_name | Specify name of vector to hold the saved the eigenvalues in workfile. |
eigvec=mat_name | Specify name of matrix to hold the save the eigenvectors in workfile. |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
Number of Component Options
fsmethod=arg (default=“simple”) | Component retention method: “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), “simple” (simple eigenvalue methods), “user” (user-specified value). Note the following: (1) If using simple methods, the minimum eigenvalue and cumulative proportions may be specified using “minigen=” and “cproport=”. (2) If setting “fsmethod=user” to provide a user-specified value, you must specify the value with “r=”. |
r=arg (default=1) | User-specified number of components to retain (for use when “fsmethod=user”). |
mineigen=arg (default=0) | Minimum eigenvalue to retain component (when “fsmethod=simple”). |
cproport=arg (default=1.0) | Cumulative proportion of eigenvalue total to attain (when “fsmethod=simple”). |
mfmethod=arg (default=“user”) | Maximum number of components used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 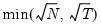 ), “size” ( 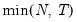 ) ), “user” (user specified value), where  is the number of series and  is the number of observations. (1) For use with all components retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of components using “rmax=”. (3) Schwert’s rule sets the maximum number of components using the rule: let for 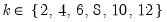 and let and let 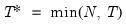 ; then the default maximum lag is given by ; then the default maximum lag is given by |
n=arg or rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Component selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Component selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Component selection (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
Eigenvalues Plot Options
The default eigenvalue graph shows a scree plot of the ordered eigenvalues. You may use the “scree”, “cproport”, and “diff” option keywords to display any combination of the scree plot, cumulative eigenvalue proportions plot, or eigenvalue difference plot.
scree | Display a scree plot of the eigenvalues (if “output=graph). |
diff | Display a graph of the eigenvalue differences (if “output=graph). |
cproport | Display a graph of the cumulative proportions (if “output=graph). |
Loadings, Scores, Biplot Graph Options
scale=arg, (default= “normload”) | Diagonal matrix scaling of the loadings and the scores: normalize loadings (“normload”), normalize scores (“normscores”), symmetric weighting (“symmetric”), user-specified (arg=number). |
cpnorm | Compute the normalization for the scores so that cross-products match the target (by default, EViews chooses a normalization scale so that the moments of the scores match the target). |
nocenter | Do not center the elements in the graph. |
mult=arg (default=”first”) | Multiple graph options: first versus remainder (“first”), pairwise (“pair”), all pairs arrayed in lower triangle (“lt”) |
labels=arg (default=“outlier”) | Scores label options: identify outliers only (“outlier”), all points (“all”), none (“none”). |
labelprob=arg (default=0.1) | Outlier label probability (if “labels=outlier”). |
autoscale=arg (default=1.0) | Rescaling factor for auto-scaling. |
userscale=arg | User-specified scaling. |
Covariance Options
cov=arg (default=“corr”) | Covariance calculation method: ordinary (Pearson product moment) covariance (“cov”), ordinary correlation (“corr”), Spearman rank covariance (“rcov”), Spearman rank correlation (“rcorr”), uncentered ordinary correlation (“ucorr”). Note that Kendall’s tau measures are not valid methods. |
wgt=name (optional) | Name of series containing weights. |
wgtmethod=arg (default = “sstdev”) | Weighting method: frequency (“freq”), inverse of variances (“var”), inverse of standard deviation (“stdev”), scaled inverse of variances (“svar”), scaled inverse of standard deviations (“sstdev”). Only applicable for ordinary (Pearson) calculations where “weights=” is specified. Weights for rank correlation and Kendall’s tau calculations are always frequency weights. |
pairwise | Compute using pairwise deletion of observations with missing cases (pairwise samples). |
partial=arg | Compute partial covariances conditioning on the list of series specified in arg. |
df | Compute covariances with a degree-of-freedom correction accounting for the mean (for centered specifications) and any partial conditioning variables. The default behavior in these cases is to perform no adjustment ( e.g. – compute sample covariance dividing by  rather than  ). |
Examples
group g1 x1 x2 x3 x4
freeze(tab1) g1.pcomp(eigval=v1, eigvec=m1)
The first line creates a group named g1 containing the four series x1, x2, x3, x4. The second line produces a view of the basic results for the principal components. The output view is stored in a table named tab1, the eigenvalues in a vector named v1, and the eigenvectors in a matrix named m1.
g1.pcomp(out=graph)
g1.pcomp(out=graph, scree, cproport)
displays a screen plot of the eigenvalues, and a graph containing both a screen plot and a plot of the cumulative eigenvalue proportions.
g1.pcomp(out=loading)
displays a loadings plot, and
g1.pcomp(out=biplot, scale=symmetric, mult=lt) 1 2 3
displays a symmetric biplot for all three pairwise comparisons.
Cross-references
See
“Principal Components” for further discussion.
To save principal components scores in series in the workfile, see
Group::makepcomp.
Reorder the members of the group.
Syntax
group_name.reorder(options) arg
The members of the group will be re-ordered based on the ordering given by arg. arg is either a list of variable names denoting the new ordering, or a keyword specifying a metric to use to compute the re-ordering:
@first | Order the group members based upon their first value (ignoring NAs). |
@firstna | Order the group members based upon their first value (treating NAs as a value). |
@last | Order the group members based upon their last value (ignoring NAs). |
@lastna | Order the group members based upon their last value (treating NAs as a value). |
@max | Order the group members based upon their maximum value. |
@mean | Order the group members based upon their mean. |
@median | Order the group members based upon their median value. |
@min | Order the group members based upon their minimum value. |
@obs | Order the group members based upon the number of non-NA observations. |
@rand | Randomize the ordering. |
@reverse | Reverse the current ordering. |
Options
Examples
group g1 v w x y z
g1.reorder x y z w v
creates a group containing V W X Y Z, then reorders the group members to X Y Z W V.
g1.reorder @mean
reorders the group members based upon their mean (highest to lowest).
g1.reorder(r) @mean
reorders the group members based upon their mean (lowest to highest).
Resample from observations in a group.
Syntax
group_name.resample(options) [output_spec]
You should follow the resample keyword with options, and if desired, an output_spec containing a list of names or a wildcard expression identifying the series to hold the output.
• If a output_spec is a list of target series, the number of names must match the number of target series.
• If you provide a wildcard output_spec, the names of the original series will be used along with the wildcard to construct the output series names. You may not use a wildcard if the series in the groups are expressions.
For example, resampling from a group containing the series X(–1) or LOG(X) will produce an error with a wildcard spec since EViews will attempt to append a suffix to the original name, producing an invalid object name.
By default, EViews uses the wildcard spec “*_b” as the output_spec.
Options
outsmpl= smpl_spec | Sample to fill the new series. Either provide the sample range in double quotes or specify a named sample object. The default is the current workfile sample. |
name= group_name | Name of group to hold created series. |
permute | Draw from rows without replacement. Default is to draw with replacement. |
weight= series_name | Name of series to be used containing values proportional to the desired row probabilities (importance sampling). The weight series must have non-missing non-negative values in the input sample, but the weights need not add up to 1, as EViews will normalize the weights. If no weights are provided, rows will be drawn with equal probability weights. |
block=integer | Block length for each draw. Must be a positive integer. The default block length is 1. |
withna (default) | [Draw / Do not draw] from all rows in the current sample, including those with NAs. |
dropna | Do not draw from rows that contain missing values in the current workfile sample. |
fixna | Excludes NAs from draws but copies rows containing missing values to the output series. |
nopanel | Ignore panel structure when resampling. By default, EViews will resample within each cross-section and assign to the corresponding cross-section. |
prompt | Force the dialog to appear from within a program. |
• If the group name specified using “name=” provide already exists and is a group object, the group object will be overwritten. If the object already exists but is not a group object, EViews will error.
• Block bootstrap (“block=” length larger than 1) requires a contiguous output sample. A block length larger than 1 cannot be used together with the “fixna” option, and the “outsmpl=” should not contain any gaps.
• The “fixna” option will have an effect only if there are missing values in the overlapping sample of the input sample (current workfile sample) and the output sample specified by “outsmpl”.
• If you specify “fixna”, we first copy any missing values in the overlapping sample to the output series. Then the input sample is adjusted to drop rows containing missing values and the output sample is adjusted so as not to overwrite the copied values.
• If you choose “dropna” and the block length is larger than 1, the input sample may shrink in order to ensure that there are no missing values in any of the drawn blocks.
• If you choose “permute”, the block option will be reset to 1, the “dropna” and “fixna” options will be ignored (reset to the default “withna” option), and the “weight” option will be ignored (reset to default equal weights).
Examples
group g1 x y
g1.resample
creates new series X_B and Y_B by drawing with replacement from the rows of X and Y in the current workfile sample. If X_B or Y_B already exist in the workfile, they will be overwritten if they are series objects, otherwise EViews will error. Note that only values of X_B and Y_B in the output sample (in this case the current workfile sample) will be overwritten.
g1.resample(weight=wt, name=G2) *_2
will append “_2” to the names for the new series, and will create a group objected named G2 containing these series. The rows in the sample will be drawn with probabilities proportional to the corresponding values in the series WT. WT must have non-missing, non-negative values in the current workfile sample.
Cross-references
See
“Resample” for a discussion of the resampling procedure. For additional discussion of wildcards, see
Appendix A. “Wildcards”.
See also
@resample and
@permute for sampling from matrices.
Set the object attribute.
Syntax
group_name.setattr(attr) attr_value
Sets the attribute attr to attr_value. Note that quoting the arguments may be required. Once added to an object, the attribute may be extracted using the @attr data member.
Examples
a.setattr(revised) never
String s = a.@attr("revised")
sets the “revised” attribute in the object A to the string “never”, and extracts the attribute into the string object S.
Cross-references
Set the fill (background) color used in the group spreadsheet using values in the spreadsheet or in a different series.
Syntax
group_name.setfillcolor(spec) fill_color_args
where the required spec is one of the following:
type = arg | Type of fill coloring for spreadsheet cells: “single” (single color), “posneg” (positive-negative single threshold), “range” (single range coloring), “hilo” (high-low-median), “custom” (custom coloring). |
s = arg | Colormap source: “none” (do not use a colormap and therefore do not color) or “series” (use the same colormap used by the individual series. |
The first form specifies a colormap for all of the series in the group. The second form, uses individual colormaps obtained from the individual series.
General Arguments
To specify the series or expression whose values will determine the background color:
• mapser(spec)
where spec is a series name or expression.
To specify the minimum and maximum values where the coloring begins and ends:
• min(color_arg)
• max(color_arg)
To set the missing value (NA) background color:
• naclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the color defaults to “white”.
Type-specific Arguments
There are optional type-specific arguments that correspond to each of the type choices:
Single color
To set the single background color:
clr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the color defaults to “white”.
Positive-negative single threshold
You may set the color for both the non-negative (posclr) and the negative (negclr) values
posclr(color_arg)
negclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the non-negative color defaults to “white” and the negative color defaults to light-red.
Single range
To specify the range, you must specify the range endpoints:
range(lower_val, upper_val[, range_def)
where range_def specifies the range endpoints:
cright | closed on the right only |
cboth | closed on both sides |
cleft | closed on the left only |
oboth | open on both sides |
By default, the range will be open on the lower and closed on the upper threshold limits.
You should provide a color specification for the inside range color (inclr) and outside range color (outclr):
inclr(color_arg)
outclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the interior color defaults to light-red, and the exterior defaults to white.
High-Low-Median
When “type=hilo” you may specify the high, low, and median coloring values:
highclr(color_arg)
lowclr(color_arg)
medianclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the colors default to light-red.
Custom
When “type=custom” you may specify custom coloring options.
You may optionally set a base background color, and then add one or more custom threshold or range color specifications. Multiple threshold and range specifications will layer, with the first applied first, followed by the second, and so on.
Custom Base Color
To set the base color (optional):
clr(color_arg)
as described below in
“Color definitions”. If omitted, the color defaults to “white”.
Custom Threshold
To add a threshold specification:
thresh(limit(threshold_value, threshold_spec), lowclr(below_arg), highclr(above_arg), threshold_name])
where threshold_spec is one of
cright | closed on the right |
cleft | closed on the left |
and the below_arg and above_arg are one of
color_arg | solid color specification |
@grad(color_arg) | gradient using color specification |
@trans | transparent |
and
color_arg are as described below in
“Color definitions”. If omitted, the color defaults to “white”.
The optional threshold_name argument may be used to attach a name to the corresponding definition.
Custom Range
To add a range specification:
range(limit(low_value, high_value, range_spec), inclr(inside_arg), outclr(outside_arg)[, range_name])
where range_spec is one of
cright | closed on the right only |
cboth | closed on both sides |
cleft | closed on the left only |
oboth | open on both sides |
inside_arg is one of
color_arg | solid color specification |
@grad(color_arg1, color_arg2) | gradient using color specification, where color_arg1 and color_arg2 are the low and high colors, respectively. |
@trans | transparent |
outside_arg is one of
color_arg | solid color specification |
@grad(color_arg) | gradient using color specification |
@trans | transparent |
color_arg1 and
color_arg2 are as described below in
“Color definitions”.
The optional range_name argument may be used to attach a name to the corresponding definition.
Color definitions
color_arg specifies the color to be employed in the arguments above. The color may be specified using predefined color names, by specifying the individual red-green-blue (RGB) components using the special “@RGB” function, or by specifying the individual red-green-blue (RGB) components in hexadecimal using the special “@HEX” function.
The predefined colors are given by the keywords (with their RGB and HEX equivalents):
blue | @rgb(0, 0, 255) | @hex(0000ff) |
red | @rgb(255, 0, 0) | @hex(ff0000) |
green | @rgb(0, 128, 0) | @hex(ffa8a8) |
black | @rgb(0, 0, 0) | @hex(008000) |
white | @rgb(255, 255, 255) | @hex(000000) |
purple | @rgb(128, 0, 128) | @hex(ffffff) |
orange | @rgb(255, 128, 0) | @hex(800080) |
yellow | @rgb(255, 255, 0) | @hex(ff8000) |
gray | @rgb(128, 128, 128) | @hex(ffff00) |
ltgray | @rgb(192, 192, 192) | @hex(808080) |
Examples
To set a gray background color for all cells in the spreadsheet, you may use:
grp1.setfillcolor(type=single) clr(gray)
To set a background color for negative values, you may use
grp1.setfillcolor(type=posneg) mapser(ser1)
which sets the background sheet fill color to white for non-negative values and light red for negative values of SER1.
Similarly,
grp1.setfillcolor(type=posneg) mapser(ser1) posclr(@rgb(10, 20, 30)) negclr(purple)
sets the background sheet fill color to @rgb(10, 20, 30) for non-negative values and purple for negative values of SER1.
Range coloring may be specified using the “type=range” option. The command
grp1.setfillcolor(type=range) mapser(ser1) clr(ltgray) range(10, 20, cleft) inclr(@rgb(128, 0, 128)) outclr(ltred) naclr(green)
sets the background fill to @rgb(128, 0, 128) for values between 10 and 20, light-red to values outside of the range 10 to 20, and green, for missing values.
Custom coloring allows you to construct more complex background filling:
grp1.setfillcolor(type=custom) mapser(ser1) clr(@rgb(10, 0, 0)) range(limit(-10, 10, oboth), inclr(green), outclr(white))) thresh(limit(-1, oleft), highclr(grey), lowclr(@trans))
Cross-references
Set the display format for cells in a group spreadsheet view.
Syntax
group_name.setformat(col_range) format_arg
where format_arg is a set of arguments used to specify format settings. If necessary, you should enclose the format_arg in double quotes.
The col_range option is used to describe the columns to be updated in groups. It may take one of the following forms:
@all | Apply to all series in the group. |
col | Column number or letter (e.g., “2”, “B”). Apply to the series corresponding to the column. |
first_col[:]last_col | Colon delimited range of columns (from low to high, e.g., “3:5”). Apply to all series corresponding to the column range. |
first_series[:]last_series | Colon delimited range of columns (from low to high, e.g., “series01:series05”) specified by the series names. Apply to all series corresponding to the column range. |
To format numeric values, you should use one of the following format specifications:
g[.precision] | significant digits |
f[.precision] | fixed decimal places |
c[.precision] | fixed characters |
e[.precision] | scientific/float |
p[.precision] | percentage |
r[.precision] | fraction |
To specify a format that groups digits into thousands using a comma separator, place a “t” after the format character. For example, to obtain a fixed number of decimal places with commas used to separate thousands, use “ft[.precision]”.
To use the period character to separate thousands and commas to denote decimal places, use “..” (two periods) when specifying the precision. For example, to obtain a fixed number of characters with a period used to separate thousands, use “ct[..precision]”.
If you wish to display negative numbers surrounded by parentheses (i.e., display the number -37.2 as “(37.2)”), you should enclose the format string in “()” (e.g., “f(.8)”).
To format numeric values using date and time formats, you may use a subset of the possible date format strings (see
“Date Formats”). The possible format arguments, along with an example of the date number 730856.944793113 (January 7, 2002 10:40:30.125 p.m) formatted using the argument are given by:
WF | (uses current EViews workfile period display format) |
YYYY | “2002” |
YYYY-Mon | “2002-Jan” |
YYYYMon | “2002 Jan” |
YYYY[M]MM | “2002[M]01” |
YYYY:MM | “2002:01” |
YYYY[Q]Q | “2002[Q]1” |
YYYY:Q | “2002:Q |
YYYY[S]S | “2002[S]1” (semi-annual) |
YYYY:S | “2002:1” |
YYYY-MM-DD | “2002-01-07” |
YYYY Mon dd | “2002 Jan 7” |
YYYY Month dd | “2002 January 7” |
YYYY-MM-DD HH:MI | “2002-01-07 22:40” |
YYYY-MM-DD HH:MI:SS | “2002-01-07 22:40:30” |
YYYY-MM-DD HH:MI:SS.SSS | “2002-01-07 22:40:30.125” |
Mon-YYYY | “Jan-2002” |
Mon dd YYYY | “Jan 7 2002” |
Mon dd, YYYY | “Jan 7, 2002” |
Month dd YYYY | “January 7 2002” |
Month dd, YYYY | “January 7, 2002” |
MM/DD/YYYY | “01/07/2002” |
mm/DD/YYYY | “1/07/2002” |
mm/DD/YYYY HH:MI | “1/07/2002 22:40” |
mm/DD/YYYY HH:MI:SS | “1/07/2002 22:40:30” |
mm/DD/YYYY HH:MI:SS.SSS | “1/07/2002 22:40:30.125” |
mm/dd/YYYY | “1/7/2002” |
mm/dd/YYYY HH:MI | “1/7/2002 22:40” |
mm/dd/YYYY HH:MI:SS | “1/7/2002 22:40:30” |
mm/dd/YYYY HH:MI:SS.SSS | “1/7/2002 22:40:30.125” |
dd/MM/YYYY | “7/01/2002” |
dd/mm/YYYY | “7/1/2002” |
DD/MM/YYYY | “07/01/2002” |
dd Mon YYYY | “7 Jan 2002” |
dd Mon, YYYY | “7 Jan, 2002” |
dd Month YYYY | “7 January 2002” |
dd Month, YYYY | “7 January, 2002” |
dd/MM/YYYY HH:MI | “7/01/2002 22:40” |
dd/MM/YYYY HH:MI:SS | “7/01/2002 22:40:30” |
dd/MM/YYYY HH:MI:SS.SSS | “7/01/2002 22:40:30.125” |
dd/mm/YYYY hh:MI | “7/1/2002 22:40” |
dd/mm/YYYY hh:MI:SS | “7/1/2002 22:40:30” |
dd/mm/YYYY hh:MI:SS.SSS | “7/1/2002 22:40:30.125” |
hm:MI am | “10:40 pm“ |
hm:MI:SS am | “10:40:30 pm” |
hm:MI:SS.SSS am | “10:40:30.125 pm” |
HH:MI | “22:40” |
HH:MI:SS | “22:40:30” |
HH:MI:SS.SSS | “22:40:30.125” |
hh:MI | “22:40” |
hh:MI:SS | “22:40:30” |
hh:MI:SS.SSS | “22:40:30.125” |
Note that the “hh” formats display 24-hour time without leading zeros. In our examples above, there is no difference between the “HH” and “hh” formats for 10 p.m.
Also note that all of the “YYYY” formats above may be displayed using two-digit year “YY” format.
Examples
To set the format for a series in a group, provide the column identifier and format:
group1.setformat(1) f.5
sets the first series in GROUP1 to fixed 5-digit precision.
group1.setformat(2) f(.7)
group1.setformat(c) e.5
sets the formats for the second and third series in the group.
You may use any of the date formats given above:
group1.setformat(2) YYYYMon
group1.setformat(d) "YYYY-MM-DD HH:MI:SS.SSS"
The column identifier may be the series names. Assuming we have a group which contains the series A1, C1, B2, A5, and H2, in that order,
group1.setformat(c1:a5) p.3
sets the formats of the series C1, B2, and A5.
Cross-references
See
Group::setwidth,
Group::setindent and
Group::setjust for details on setting spreadsheet widths, indentation and justification.
Set the display indentation for cells in a group object spreadsheet view.
Syntax
group_name.setindent(col_range) indent_arg
where indent_arg is an indent value specified in 1/5 of a width unit. The width unit is computed from representative characters in the default font for the current spreadsheet (the EViews spreadsheet default font at the time the spreadsheet was created), and corresponds roughly to a single character. Indentation is only relevant for non-center justified cells.
The default indentation setttings are taken from the Global Defaults for spreadsheet views (
“Spreadsheet Data Display”) at the time the spreadsheet was created.
The
col_range option is used to describe the columns to be updated. See
Group::setformat for the syntax for
col_range specifications.
Examples
To set the justification, provide the column identifier and the format. The commands,
group1.setindent(2) 3
group1.setindent(c) 2
set the formats for the second and third series in the group, while:
group2.setindent(@all) 3
sets formats for all of the series.
Cross-references
See
Group::setwidth and
Group::setjust for details on setting spreadsheet widths and justification.
Set the justification for cells in the spreadsheet view of the group object.
Syntax
group_name.setjust(col_range) format_arg
where format_arg may consist of the following:
top / middle / bottom | Vertical justification setting. |
auto / left / center / right | Horizontal justification setting. Strings are left-justified and numbers are right-justified under “auto”. |
The
col_range option is used to specify the columns to be justified in the group. See
Group::setformat for syntax.
Default display settings can be set in General Options; see
“Spreadsheet Data Display”.
Examples
To set the justification, provide the column identifier and the format. The commands
group1.setjust(2) bottom center
group1.setjust(c) center middle
set the formats for the second and third series in the group GROUP1.
group2.setjust(@all) right
sets the format for all series in the group GROUP2.
Cross-references
See
Group::setwidth and
Group::setindent for details on setting spreadsheet widths and indentation.
Set the text color used in the group spreadsheet using values in the spreadsheet or in a different series.
Syntax
group_name.settextcolor(spec) fill_color_args
where the required spec is one of the following:
type = arg | Type of fill coloring for spreadsheet cells: “single” (single color), “posneg” (positive-negative single threshold), “range” (single range coloring), “hilo” (high-low-median), “custom” (custom coloring). |
s = arg | Colormap source: “none” (do not use a colormap and therefore do not color) or “series” (use the same colormap used by the individual series. |
The first form specifies a colormap for all of the series in the group. The second form, uses individual colormaps obtained from the individual series.
General Arguments
To specify the series or expression whose values will determine the background color:
• mapser(spec)
where spec is a series name or expression.
To specify the minimum and maximum values where the coloring begins and ends:
• min(color_arg)
• max(color_arg)
To set the missing value (NA) background color:
• naclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the color defaults to “black”.
Type-specific Arguments
There are optional type-specific arguments that correspond to each of the type choices:
Single color
To set the single text color:
clr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the color defaults to “black”.
Positive-negative single threshold
You may set the color for both the non-negative (posclr) and the negative (negclr) values
posclr(color_arg)
negclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the non-negative color defaults to “black” and the negative color defaults to “red”.
Single range
To specify the range, you must specify the range endpoints:
range(lower_val, upper_val[, range_def)
where range_def specifies the range endpoints:
cright | closed on the right only |
cboth | closed on both sides |
cleft | closed on the left only |
oboth | open on both sides |
By default, the range will be open on the lower and closed on the upper threshold limits.
You should provide a color specification for the inside range color (inclr) and outside range color (outclr):
inclr(color_arg)
outclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the interior color defaults to light-red, and the exterior defaults to white.
High-Low-Median
When “type=hilo” you may specify the high, low, and median coloring values:
highclr(color_arg)
lowclr(color_arg)
medianclr(color_arg)
where
color_arg is described below in
“Color definitions”. If omitted, the colors default to light-red.
Custom
When “type=custom” you may specify custom coloring options.
You may optionally set a base text color, and then add one or more custom threshold or range color specifications. Multiple threshold and range specifications will layer, with the first applied first, followed by the second, and so on.
Custom Base Color
To set the base color (optional):
clr(color_arg)
as described below in
“Color definitions”. If omitted, the color defaults to “white”.
Custom Threshold
To add a threshold specification:
thresh(limit(threshold_value, threshold_spec), lowclr(below_arg), highclr(above_arg), threshold_name])
where threshold_spec is one of
cright | closed on the right |
cleft | closed on the left |
and the below_arg and above_arg are one of
color_arg | solid color specification |
@grad(color_arg) | gradient using color specification |
@trans | transparent |
and
color_arg are as described below in
“Color definitions”. If omitted, the color defaults to “white”.
The optional threshold_name argument may be used to attach a name to the corresponding definition.
Custom Range
To add a range specification:
range(limit(low_value, high_value, range_spec), inclr(inside_arg), outclr(outside_arg)[, range_name])
where range_spec is one of
cright | closed on the right only |
cboth | closed on both sides |
cleft | closed on the left only |
oboth | open on both sides |
inside_arg is one of
color_arg | solid color specification |
@grad(color_arg1, color_arg2) | gradient using color specification, where color_arg1 and color_arg2 are the low and high colors, respectively. |
@trans | transparent |
outside_arg is one of
color_arg | solid color specification |
@grad(color_arg) | gradient using color specification |
@trans | transparent |
color_arg1 and
color_arg2 are as described below in
“Color definitions”.
The optional range_name argument may be used to attach a name to the corresponding definition.
Color definitions
color_arg specifies the color to be employed in the arguments above. The color may be specified using predefined color names, by specifying the individual red-green-blue (RGB) components using the special “@RGB” function, or by specifying the individual red-green-blue (RGB) components in hexadecimal using the special “@HEX” function.
The predefined colors are given by the keywords (with their RGB and HEX equivalents):
blue | @rgb(0, 0, 255) | @hex(0000ff) |
red | @rgb(255, 0, 0) | @hex(ff0000) |
ltred | @rgb(255, 168, 168) | @hex(ffa8a8) |
green | @rgb(0, 128, 0) | @hex(008000) |
black | @rgb(0, 0, 0) | @hex(000000) |
white | @rgb(255, 255, 255) | @hex(ffffff) |
purple | @rgb(128, 0, 128) | @hex(800080) |
orange | @rgb(255, 128, 0) | @hex(ff8000) |
yellow | @rgb(255, 255, 0) | @hex(ffff00) |
gray | @rgb(128, 128, 128) | @hex(808080) |
ltgray | @rgb(192, 192, 192) | @hex(c0c0c0) |
Examples
To set a gray text color for all cells in the spreadsheet, you may use:
grp1.settextcolor(type=single) clr(gray)
To set a text color for negative values, you may use
grp1.settextcolor(type=posneg) mapser(ser1)
which sets the text color to black for non-negative values and red for negative values of SER1.
Similarly,
grp1.settextcolor(type=posneg) mapser(ser1) posclr(@rgb(10, 20, 30)) negclr(purple)
sets the text color to @rgb(10, 20, 30) for non-negative values and purple for negative values of SER1.
Range coloring may be specified using the “type=range” option. The command
grp1.settextcolor(type=range) mapser(ser1) clr(ltgray) range(10, 20, cleft) inclr(@rgb(128, 0, 128)) outclr(ltred) naclr(green)
sets the text to @rgb(128, 0, 128) for values between 10 and 20, light-red to values outside of the range 10 to 20, and green, for missing values.
Custom coloring allows you to construct more complex text coloring:
grp1.settextcolor(type=custom) mapser(ser1) clr(@rgb(10, 0, 0)) range(limit(-10, 10, oboth), inclr(green), outclr(white))) thresh(limit(-1, oleft), highclr(grey), lowclr(@trans))
Cross-reference
Set the column width for selected columns in a group spreadsheet.
Syntax
group_name.setwidth(col_range) width_arg
where col_range is either a single column number or letter (e.g., “5”, “E”), a colon delimited range of columns (from low to high, e.g., “3:5”, “C:E”), or the keyword “@ALL”, and width_arg specifies the width unit value. The width unit is computed from representative characters in the default font for the current spreadsheet (the EViews spreadsheet default font at the time the spreadsheet was created), and corresponds roughly to a single character. width_arg values may be non-integer values with resolution up to 1/10 of a width unit.
Examples
gr1.setwidth(2) 12
sets the width of column 2 to 12 width units.
gr1.setwidth(2:10) 20
sets the widths for columns 2 through 10 to 20 width units.
Cross-references
See
Group::setindent and
Group::setjust for details on setting spreadsheet indentation and justification.
Spreadsheet view of a group object.
Syntax
group_name.sheet(options)
Options
w | Wide. In a panel this will switch to the unstacked form of the panel (dates along the side, cross-sections along the top). |
t | Transpose. |
a | All observations (ignore sample) |
nl | Do not display labels. |
tform=arg (default= “level” | Display transformed data: raw data (“level”), one period difference (“dif” or “d”), annual difference (“dify” or “dy”), one period percentage change (“pch” or “pc”), annualized one period percentage change (“pcha” or “pca”), annual percentage change (“pchy” or “pcy”), natural logarithm (“log”), one period difference of logged values (“dlog”). |
c | Compare view. Display the compare view of the group. |
p | Print the spreadsheet view. |
Examples
g1.sheet(p)
displays and prints the spreadsheet view of the group G1.
g1.sheet(t, tform=log)
shows log values of the series in G1 using the current sample in a wide spreadsheet.
g1.sheet(nl, tform=diff)
displays differenced values of the series in the group using the current sample with no labels.
g1.sheet(a, tform=pc)
displays the one period percent changes for all observations in the workfile.
Cross-references
See
“Basic Data Handling” for a discussion of the spreadsheet view of series and groups.
Change display order for group spreadsheet.
The sort command changes the sort order settings for spreadsheet display of the group.
Syntax
group_name.sort(series1[, series2, series3])
Follow the keyword with a list of the series you wish to use to determine display order. You may specify up to three series for sorting. If you list two or more series, sort uses the values of the second series to resolve ties in the first series, and values of the third series to resolve ties in the first and second. By default, EViews will sort in ascending order. For purposes of sorting, NAs are considered to be smaller than any other value.
The series may be specified using the name or index of a series in the group. For example, if you provide the integer “2”, EViews will use the second series. To sort by the original workfile observation order, use the integer “0”, or the keyword “obs”.
To sort in descending order, precede the series name or index with a minus sign (“-”).
Examples
gr1.sort(x,y)
change the display order for group GR1, sorting by the series X and Y, with ties in X resolved using Y.
If X is the first series in group GR1 and Y is the second series,
gr1.sort(1,-2)
sorts first in ascending order by X and then in descending order by Y.
gr1.sort(obs)
returns the display order for group GR1 to the original (by observation).
Cross-references
See
“Spreadsheet” for additional discussion.
Descriptive statistics for series in a group.
Computes and displays a table of means, medians, maximum and minimum values, standard deviations, and other descriptive statistics for each series in the group.
Syntax
group_name.stats(options)
Options
i | Use individual sample for each series after removing missing values for the individual series. By default, EViews computes the statistics using a common sample after removing observations with missing values for any series in the group. |
p | Print the stats table. |
Examples
The commands
group group1 wage hrs edu
group01.stats
computes the descriptive statistics for each series in GROUP01 for the balanced sample obtained after deletion of observation in the current sample with missing values for any of the series Alternately,
group01.stats(i)
displays the descriptive statistics view of GROUP01 showing the statistics for each series computed using individual samples.
Cross-references
See
“Descriptive Statistics” for a discussion of the descriptive statistics views of a group.
Test equality of the mean, median or variance between (among) series in a group.
Syntax
group_name.testbtw(options)
Specify the type of test as an option.
Options
mean (default) | Test equality of mean. |
med | Test equality of median. |
var | Test equality of variance. |
c | Use common sample. |
i (default) | Use individual sample. |
prompt | Force the dialog to appear from within a program. |
p | Print the test results. |
Examples
group g1 wage_m wage_f
g1.testbtw
g1.testbtw(var,c)
tests the equality of means between the two series WAGE_M and WAGE_F.
Cross-references
See
“Tests of Equality” for further discussion of these tests.
Carries out (panel) unit root tests on a group of series.
When used on a group of series, the procedure will perform panel unit root testing. The panel unit root tests include Levin, Lin and Chu (LLC), Breitung, Im, Pesaran, and Shin (IPS), Fisher - ADF, Fisher - PP, and Hadri tests on levels, or first or second differences.
Syntax
group_name.uroot(options)
Options
Basic Specification Options
You should specify the exogenous variables and order of dependent variable differencing in the test equation using the following options:
const (default) | Include a constant in the test equation. |
trend | Include a constant and a linear time trend in the test equation. |
none | Do not include a constant or time trend (only available for the ADF and PP tests). |
dif=integer (default=0) | Order of differencing of the series prior to running the test. Valid values are {0, 1, 2}. |
You may use one of the following keywords to specify the test:
sum (default) | Summary of the first five panel unit root tests (where applicable). |
llc | Levin, Lin, and Chu. |
breit | Breitung. |
ips | Im, Pesaran, and Shin. |
adf | Fisher - ADF. |
pp | Fisher - PP. |
hadri | Hadri. |
Sample Option
balance | Use balanced (across cross-sections or series) data when performing test. |
Lag Difference Options
Specifies the number of lag difference terms to be included in the test equation. Applicable in “Summary”, LLC, Breitung, IPS, and Fisher-ADF tests. The default setting depends on whether you choose to balance the samples across cross-sections.
If you do not include the “balance” option, the default is to perform automatic lag selection using the Schwarz criteria (“lagmethod=sic”).
Alternately, if you include the “balance” option, the default setting is a common, observation-based fixed lag (“lag=default”) where:
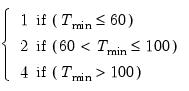 | (1.1) |
lagmethod=arg (default=“sic”) | Method for selecting lag lengths (number of first difference terms) to be included in the Dickey-Fuller test regressions: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “tstat” (Ng-Perron first backward significant t-statistic). |
lag=arg | Specified lag length (number of first difference terms) to be included in the regression: integer (user-specified common lag length), vector_name (user-specific individual lag length, one row per cross-section). |
maxlag=arg | Maximum lag length to consider when performing automatic lag length selection: integer (common maximum lag length), or vector_name (individual maximum lag length, one row per cross-section). The default setting produces individual maximum lags of, default= 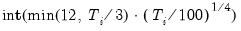 where  is the length of the cross-section. |
lagpval=arg (default=0.1) | Probability value for use in the t-statistic automatic lag selection method (when “lagmethod = tstat”). |
Kernel Options
Specifies options for computing kernel estimates of the zero-frequency spectrum (long-run covariance). Applicable to “Summary”, LLC, Fisher-PP, and Hadri tests.
hac=arg (default=“bt”) | Method of estimating the frequency zero spectrum: “bt” (Bartlett kernel), “pr” (Parzen kernel), “qs” (Quadratic Spectral kernel), |
band = arg, b=arg (default=“nw”) | Method of selecting the bandwidth: “nw” (Newey-West automatic variable bandwidth selection), “a” (Andrews automatic selection), number (user-specified common bandwidth), vector_name (user-specified individual bandwidths, one row for each cross-section). |
Other options
prompt | Force the dialog to appear from within a program. |
p | Print output from the test. |
Examples
The command:
Grp1.uroot(llc,exog=trend)
performs the LLC panel unit root test with exogenous individual trends and individual effects on series in GRP1.
Grp2.uroot(is,exog=const,maxlag=4,lagmethod=AIC)
performs the IPS panel unit root test on series in group GP2. The test includes individual effects, lag will be chosen by AIC from maximum lag of three.
Grp3.uroot(sum,exog=const,lag=3,hac=pr,b=2.3)
performs a summary of the panel unit root tests on the series in group GP3. The test equation includes a constant term and three lagged first-difference terms. The frequency zero spectrum is estimated using kernel methods (with a Parzen kernel), and a bandwidth of 2.3.
Cross-references
See
“Unit Root Testing” for discussion of standard unit root tests performed on a single series, and
“Cross-sectionally Independent Panel Unit Root Testing” and
“Cross-sectionally Dependent Panel Unit Root Tests” for discussion of unit roots tests performed on panel structured workfiles, groups of series, or pooled data.
Compute dependent (second generation) panel unit root tests on a group of series.
Syntax
group_name.uroot2(options)
where group_name is the name of a group object.
Options
General Options
type=arg (default=“panic”) | Type of unit root test: PANIC - Bai and Ng (2004) (“panic”), CIPS - Pesaran (2007) (“cips”). Note: (1) when performing PANIC testing, factor selection, MQ, ADF lag selection, VAR lag selection (possibly), long-run variance (possibly), and p-value simulation options are relevant. (2) when perform CIPS testing, ADF lag selection options are relevant. |
exog=arg (default=“constant”) | Exogenous deterministic variables to include for each cross-section: “none” (no deterministic variables), “constant” (only a constant), “trend” (both a constant and trend). |
prompt | Force the dialog to appear from within a program. |
p | Print results. |
ADF Lag Selection Options
adflagmethod=arg (default=“sic”) | Method for selecting lag length (number of first difference terms) to be included in the Dickey-Fuller test regression or number of lags in the AR spectral density estimator: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “msaic” (Modified Akaike), “msic” (Modified Schwarz), “mhqc” (Modified Hannan-Quinn), “tstat” (Ng-Perron first backward significant t-statistic). |
adflag=integer | Use-specified fixed lag. |
adfmaxlag=integer | Maximum lag length to consider when performing automatic lag length selection. Note: default is Schwert’s rule: let for  and let and let 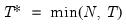 ; then the default maximum lag is given by ; then the default maximum lag is given by |
adflagpval=arg (default=0.1) | Probability value for use in the t-statistic automatic lag selection method (“lagmethod = tstat”). |
PANIC Number of Factor Selection Options
fsmethod=arg (default=“bn”) | Factor retention selection method: “bn” (Bai and Ng (2002)), “ah” (Ahn and Horenstein (2013)), “simple” (simple eigenvalue methods), “user” (user-specified value). Note the following: (1) If using simple methods, the minimum eigenvalue and cumulative proportions may be specified using “minigen=” and “cproport=”. (2) If setting “fsmethod=user” to provide a user-specified value, you must specify the value with “r=”. |
r=arg (default=1) | User-specified number of factors to retain (for use when “fsmethod=user”). |
mineigen=arg (default=0) | Minimum eigenvalue to retain factor (when “fsmethod=simple”). |
cproport=arg (default=1.0) | Cumulative proportion of eigenvalue total to attain (when “fsmethod=simple”). |
mfmethod=arg | Maximum number of factors used by selection methods: “schwert” (Schwert’s rule, default), “ah” (Ahn and Horenstein’s (2013) suggestion), “rootsize” ( 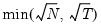 ), “size” ( 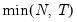 ) ), “user” (user specified value). (1) For use with all factor retention methods apart from user-specified (“fsmethod=user”). (2) If setting “mfmethod=user”, you may specify the maximum number of factors using “rmax=”. (3) Schwert’s rule sets the maximum number of factors using the rule: let for 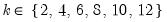 and let and let 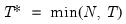 ; then the default maximum lag is given by ; then the default maximum lag is given by |
rmax=arg (default=all) | User-specified maximum number of factors to retain (for use when “mfmethod=user”). |
fsic=arg (default=avg) | Factor selection criterion (when “fsmethod=bn”): “icp1” (ICP1), “icp2” (ICP2), “icp3” (ICP3), “pcp1” (PCP1), “pcp2” (PCP1), “pcp3” (ICP3), “avg” (average of all criteria ICP1 through PCP3). Factor selection criterion (when “fsmethod=ah”): “er” (eigenvalue ratio), “gr” (growth ratio), “avg” (average of eigenvalue ratio and growth ratio). Factor selection criterion (when “fsmethod=simple”): “min” (minimum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “max” (maximum of: minimum eigenvalue, cumulative eigenvalue proportion, and maximum number of factors), “avg” (average the optimal number of factors as specified by the min and max rule, then round to the nearest integer). |
demeantime | Demeans observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizetime | Standardizes observations across time prior to component selection procedures, when “n=bn” or “n=ah”. |
demeancross | Demeans observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
sdizecross | Standardizes observations across cross-sections prior to component selection procedures, when “n=bn” or “n=ah”. |
PANIC VAR Lag Selection Options
For use when computing a PANIC test with
statistic.
varlagmethod=arg (default=“sic”) | Method for selecting lag length (number of first difference terms) to be included in the test statistic VAR: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn), “msaic” (Modified Akaike), “msic” (Modified Schwarz), “mhqc” (Modified Hannan-Quinn), “tstat” (Ng-Perron first backward significant t-statistic). |
varlag=integer | Use-specified fixed lag. |
varmaxlag=integer | Maximum lag length to consider when performing automatic lag length selection. Note: default is Schwert’s rule: let for 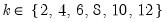 and let and let 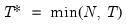 ; then the default maximum lag is given by ; then the default maximum lag is given by |
PANIC Long-run Variance Options
For use when computing a PANIC test using the
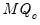
statistic.
Whitening Options
lag=arg | Lag specification: integer (user-specified number of lags), “a” (automatic selection). |
infosel=arg (default=“aic”) | Information criterion for automatic selection: “aic” (Akaike), “sic” (Schwarz), “hqc” (Hannan-Quinn) (if “lag=a”). |
maxlag=integer | Maximum lag-length for automatic selection (optional) (if “lag=a”). The default is an observation-based maximum of 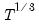 . |
Kernel Options
kern=arg (default=“bart”) | Kernel shape: “none” (no kernel), “bart” (Bartlett), “bohman” (Bohman), “daniell” (Daniel), “parzen” (Parzen), “parzriesz” (Parzen-Riesz), “parzgeo” (Parzen-Geometric), “parzcauchy” (Parzen-Cauchy), “quadspec” (Quadratic Spectral), “trunc” (Truncated), “thamm” (Tukey-Hamming), “thann” (Tukey-Hanning), “tparz” (Tukey-Parzen), “user” (User-specified; see “kernwgt=” below). |
kernwgt=vector | User-specified kernel weight vector (if “kern=user”). |
bw=arg (default=”nwfixed”) | Bandwidth: “fixednw” (Newey-West fixed), “andrews” (Andrews automatic), “neweywest” (Newey-West automatic), number (User-specified bandwidth). |
nwlag=integer | Newey-West lag-selection parameter for use in nonparametric bandwidth selection (if “bw=neweywest”). |
bwoffset=integer (default=0) | Apply integer offset to bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
bwint | Use integer portion of bandwidth chosen by automatic selection method (“bw=andrews” or “bw=neweywest”). |
PANIC p-value Options
mcreps=integer | Number of Monte Carlo replications. |
asymplen=integer | Asymptotic length of series. |
seed=number | Specifies the random number generator seed |
rng=arg | Specifies the type of random number generator. The key can be; improved Knuth generator (“kn”), improved Mersenne Twister (“mt”), Knuth’s (1997) lagged Fibonacci generator used in EViews 4 (“kn4”) L’Ecuyer’s (1999) combined multiple, recursive generator (“le”), Matsumoto and Nishimura’s (1998) Mersenne Twister used in EViews 4 (“mt4”). |
Examples
grp.uroot2
The line above performs a PANIC unit root test on the series in the group GRP.
grp.uroot2(fsmethod=AH, mq=mqf, varlag=3)
The line above performs a PANIC unit root test using Ahn and Horenstein (2013) for factor selection determination and the
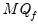
test for the number of common trends using a VAR(3) model.
grp.uroot2(test=cips, exog=trend, adnfosel=sic)
The line above performs a CIPS unit root test on the series in group GRP, with ADF testing performed on each cross-section with a constant and trend, and ADF lag selection using the Schwarz criterion.
Cross-references
See
“Unit Root Testing” for discussion of standard unit root tests performed on a single series, and
“Cross-sectionally Independent Panel Unit Root Testing” and
“Cross-sectionally Dependent Panel Unit Root Tests” for discussion of unit roots tests performed on panel structured workfiles, groups of series, or pooled data.
References
MacKinnon, James G., Alfred A. Haug, and Leo Michelis (1999), “Numerical Distribution Functions of Likelihood Ratio Tests For Cointegration,” Journal of Applied Econometrics, 14, 563-577.
Osterwald-Lenum, Michael (1992). “A Note with Quantiles of the Asymptotic Distribution of the Maximum Likelihood Cointegration Rank Test Statistics,” Oxford Bulletin of Economics and Statistics, 54, 461–472.
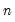 -way contingency table.
-way contingency table.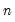 -way contingency table.
-way contingency table.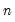 -way contingency table.
-way contingency table.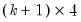 matrix, where
matrix, where 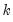 is the number of endogenous variables in the VAR. The first column contains the eigenvalues, the second column contains the maximum eigenvalue statistics, the third column contains the trace statistics, and the fourth column contains the log likelihood values. The i-th row of columns 2 and 3 are the test statistics for rank
is the number of endogenous variables in the VAR. The first column contains the eigenvalues, the second column contains the maximum eigenvalue statistics, the third column contains the trace statistics, and the fourth column contains the log likelihood values. The i-th row of columns 2 and 3 are the test statistics for rank 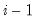 . The last row is filled with NAs, except the last column which contains the log likelihood value of the unrestricted (full rank) model.
. The last row is filled with NAs, except the last column which contains the log likelihood value of the unrestricted (full rank) model. 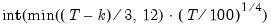
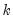 is the number of coefficients in the cointegrating equation. Applicable when “lag=a”.
is the number of coefficients in the cointegrating equation. Applicable when “lag=a”.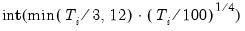
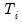 is the length of the cross-section.
is the length of the cross-section.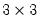 Pearson correlation matrix for the three series in GRP1.
Pearson correlation matrix for the three series in GRP1.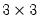 Pearson covariance matrix for the three series in GRP1.
Pearson covariance matrix for the three series in GRP1.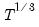 .
.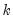 components corresponding to the
components corresponding to the 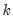 elements in output_list, up to the total number of series in the group.
elements in output_list, up to the total number of series in the group.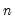 rather than
rather than 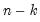 ).
).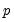 ) for the series in the group, compute the residuals, and save the results into whitened series.
) for the series in the group, compute the residuals, and save the results into whitened series.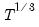 .
.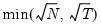 ), “size” (
), “size” (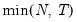 ), “user” (user specified value), where
), “user” (user specified value), where 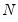 is the number of series and
is the number of series and 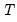 is the number of observations.
is the number of observations.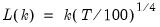
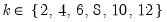 and let
and let 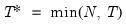 ; then the default maximum lag is given by
; then the default maximum lag is given by 
 rather than
rather than  ).
).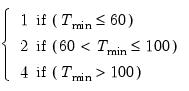
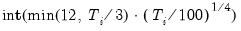
 is the length of the cross-section.
is the length of the cross-section.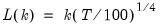
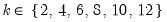 and let
and let 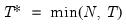 ; then the default maximum lag is given by
; then the default maximum lag is given by 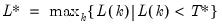
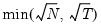 ), “size” (
), “size” (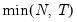 ), “user” (user specified value).
), “user” (user specified value).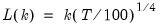
 and let
and let  ; then the default maximum lag is given by
; then the default maximum lag is given by 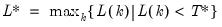
 statistic.
statistic.
 and let
and let 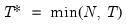 ; then the default maximum lag is given by
; then the default maximum lag is given by 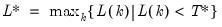
 statistic.
statistic.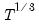 .
.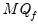 test for the number of common trends using a VAR(3) model.
test for the number of common trends using a VAR(3) model.